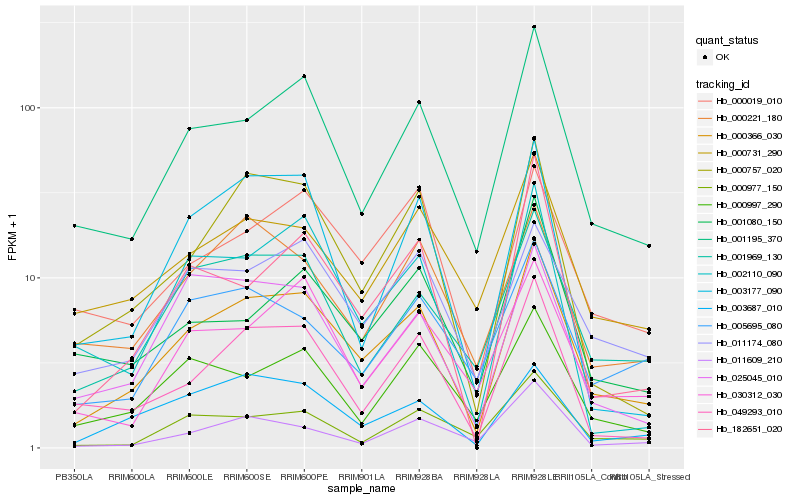
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000366_030 |
0.0 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 2 |
Hb_001195_370 |
0.1067690397 |
- |
- |
- |
| 3 |
Hb_000757_020 |
0.1282378372 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_001969_130 |
0.1329786386 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 5 |
Hb_049293_010 |
0.1349688122 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000731_290 |
0.1429785289 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_011174_080 |
0.143030338 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 8 |
Hb_003687_010 |
0.1442683262 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 9 |
Hb_000019_010 |
0.1473156247 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |
| 10 |
Hb_182651_020 |
0.1492114893 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_030312_030 |
0.1493874581 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000997_290 |
0.1504267917 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 13 |
Hb_025045_010 |
0.1513620286 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_002110_090 |
0.1527718996 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 15 |
Hb_005695_080 |
0.1544719343 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 16 |
Hb_000977_150 |
0.1554716633 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 17 |
Hb_001080_150 |
0.1555545393 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 18 |
Hb_003177_090 |
0.1570428329 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 19 |
Hb_000221_180 |
0.1571397317 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 20 |
Hb_011609_210 |
0.1611965454 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |