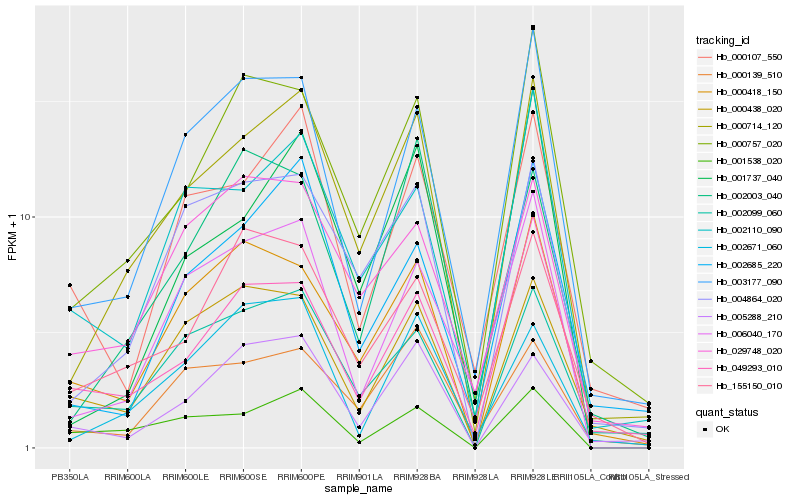
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000714_120 |
0.0 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 2 |
Hb_004864_020 |
0.1074692542 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 3 |
Hb_000757_020 |
0.1186460447 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_003177_090 |
0.1327270006 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 5 |
Hb_002099_060 |
0.1337440948 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_002110_090 |
0.1415660987 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 7 |
Hb_000418_150 |
0.1422731574 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_002685_220 |
0.142287928 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001737_040 |
0.1452251424 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 10 |
Hb_006040_170 |
0.1493238168 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 11 |
Hb_000139_510 |
0.1528674595 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_029748_020 |
0.153351072 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002671_060 |
0.1558112648 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 14 |
Hb_155150_010 |
0.1568335102 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_049293_010 |
0.157339442 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_005288_210 |
0.1576195217 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 17 |
Hb_000438_020 |
0.1578600621 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 18 |
Hb_002003_040 |
0.1589289013 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000107_550 |
0.1608621463 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 20 |
Hb_001538_020 |
0.1624909514 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |