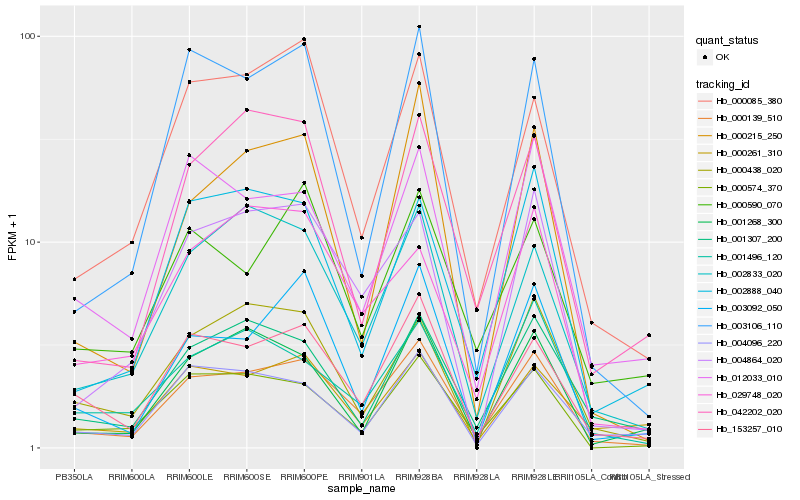
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_510 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_153257_010 |
0.107614761 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 3 |
Hb_002833_020 |
0.1208713807 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 4 |
Hb_000574_370 |
0.1252910023 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 5 |
Hb_004864_020 |
0.1278562212 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 6 |
Hb_003092_050 |
0.1313490612 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 7 |
Hb_000085_380 |
0.1314452162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001307_200 |
0.1325826732 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 9 |
Hb_001496_120 |
0.1344208438 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_012033_010 |
0.1353013677 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 11 |
Hb_002888_040 |
0.1375501049 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 12 |
Hb_003106_110 |
0.1382657799 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000438_020 |
0.1420827395 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 14 |
Hb_004096_220 |
0.1424412207 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000261_310 |
0.142653913 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 16 |
Hb_001268_300 |
0.1459715852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000590_070 |
0.1463820294 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 18 |
Hb_042202_020 |
0.1474538992 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000215_250 |
0.147735412 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_029748_020 |
0.147762493 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |