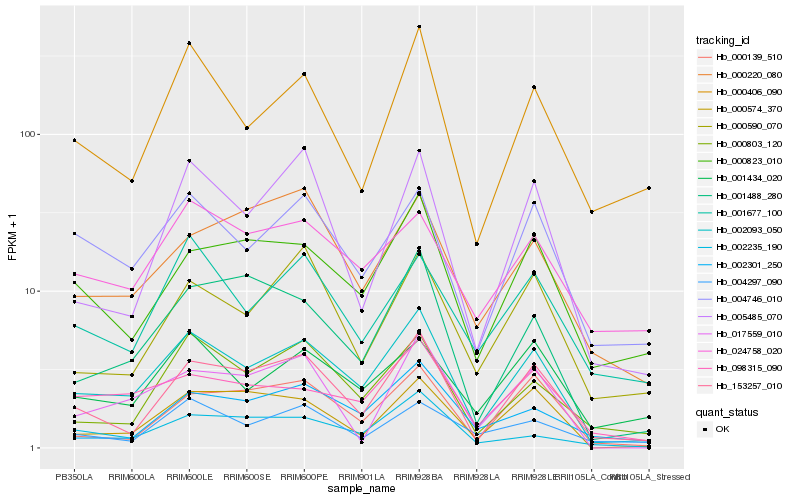
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002093_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 2 |
Hb_153257_010 |
0.1196640547 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 3 |
Hb_004746_010 |
0.1339887297 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 4 |
Hb_000406_090 |
0.1419632956 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_098315_090 |
0.1436791236 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 6 |
Hb_000823_010 |
0.1444641617 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000803_120 |
0.1450012502 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001488_280 |
0.1463197357 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 9 |
Hb_000574_370 |
0.151846673 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 10 |
Hb_001434_020 |
0.1539600936 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 11 |
Hb_005485_070 |
0.1564338361 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001677_100 |
0.1581849205 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 13 |
Hb_000590_070 |
0.1592754057 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 14 |
Hb_002235_190 |
0.1603882059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000139_510 |
0.1619309588 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_024758_020 |
0.1647230546 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 17 |
Hb_002301_250 |
0.1667675507 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a [Jatropha curcas] |
| 18 |
Hb_004297_090 |
0.1698766402 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 19 |
Hb_000220_080 |
0.1705330126 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_017559_010 |
0.1723532121 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |