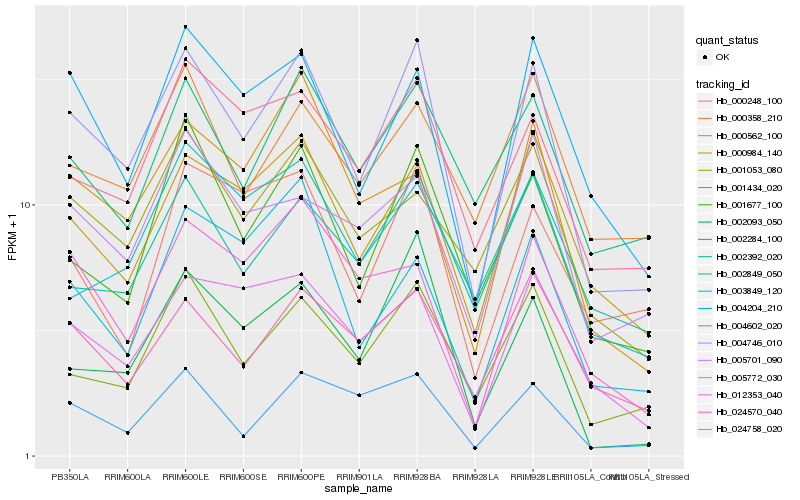
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004746_010 |
0.0 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 2 |
Hb_000984_140 |
0.1125403222 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 3 |
Hb_001434_020 |
0.1125941244 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 4 |
Hb_004204_210 |
0.1144271387 |
- |
- |
PREDICTED: heparanase-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001677_100 |
0.1183285313 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 6 |
Hb_024758_020 |
0.1229248362 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 7 |
Hb_005772_030 |
0.1249829685 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 8 |
Hb_024570_040 |
0.1288605323 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 9 |
Hb_002284_100 |
0.1319383214 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 10 |
Hb_005701_090 |
0.1332916536 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002093_050 |
0.1339887297 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 12 |
Hb_001053_080 |
0.1356367023 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 13 |
Hb_004602_020 |
0.1370275246 |
transcription factor |
TF Family: SET |
PREDICTED: N-lysine methyltransferase setd6 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002392_020 |
0.1404819068 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 15 |
Hb_000358_210 |
0.1407674265 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 16 |
Hb_000562_100 |
0.142647194 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 17 |
Hb_012353_040 |
0.1438298003 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_003849_120 |
0.147259189 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 19 |
Hb_002849_050 |
0.148195781 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 20 |
Hb_000248_100 |
0.1483620045 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |