| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
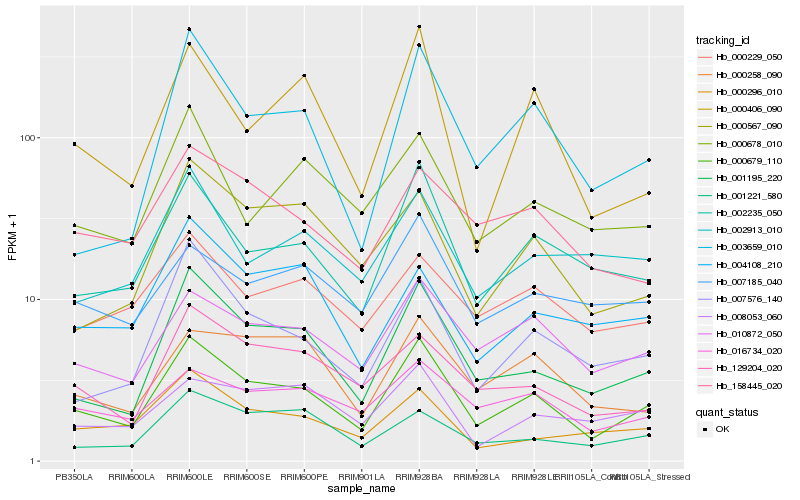
Hb_000679_110 |
0.0 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 2 |
Hb_001195_220 |
0.1048589655 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_002235_050 |
0.1156795642 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_010872_050 |
0.1260336477 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 5 |
Hb_007576_140 |
0.136180747 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 6 |
Hb_129204_020 |
0.1438634252 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 7 |
Hb_003659_010 |
0.1454639585 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 8 |
Hb_016734_020 |
0.1465467024 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 9 |
Hb_000406_090 |
0.1472983357 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_000296_010 |
0.1482540253 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 11 |
Hb_158445_020 |
0.1493113994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000567_090 |
0.149645936 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_001221_580 |
0.1500249909 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 14 |
Hb_000678_010 |
0.1514841649 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000258_090 |
0.1523883719 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007185_040 |
0.1529249213 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 17 |
Hb_000229_050 |
0.1549074623 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 18 |
Hb_002913_010 |
0.1550666015 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004108_210 |
0.1569215953 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 20 |
Hb_008053_060 |
0.1576231302 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |