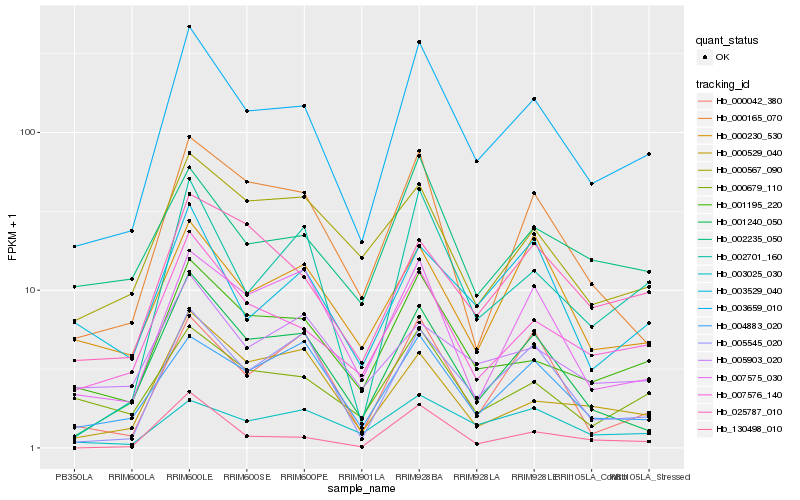
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003659_010 |
0.0 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 2 |
Hb_001195_220 |
0.1158750962 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_007576_140 |
0.1173459716 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 4 |
Hb_002701_160 |
0.1372485269 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |
| 5 |
Hb_001240_050 |
0.1416935116 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 6 |
Hb_000679_110 |
0.1454639585 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 7 |
Hb_002235_050 |
0.1454834615 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005545_020 |
0.1500106814 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 9 |
Hb_004883_020 |
0.1584912173 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 10 |
Hb_000165_070 |
0.1588859276 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 11 |
Hb_025787_010 |
0.1620979965 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 12 |
Hb_003025_030 |
0.1631497651 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 13 |
Hb_130498_010 |
0.163496412 |
- |
- |
Laccase 17 [Theobroma cacao] |
| 14 |
Hb_007575_030 |
0.163844473 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 15 |
Hb_000567_090 |
0.1685815393 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_003529_040 |
0.1695622111 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000042_380 |
0.1715775024 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 18 |
Hb_000230_530 |
0.1739402214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000529_040 |
0.1745425763 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 20 |
Hb_005903_020 |
0.1777928221 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |