| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
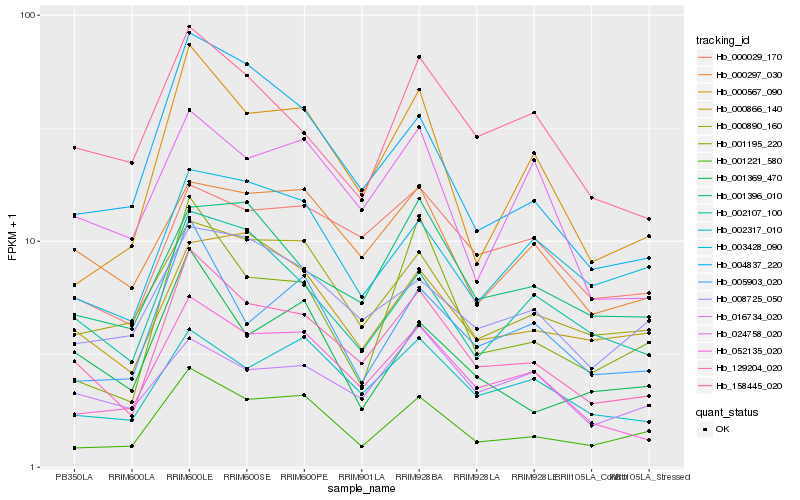
Hb_129204_020 |
0.0 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 2 |
Hb_052135_020 |
0.1006657995 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 3 |
Hb_001221_580 |
0.118064363 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 4 |
Hb_004837_220 |
0.1232250619 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 5 |
Hb_002317_010 |
0.1238002665 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_002107_100 |
0.1241474208 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_158445_020 |
0.1248129733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000567_090 |
0.1254042012 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_005903_020 |
0.1301923017 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 10 |
Hb_000890_160 |
0.1303147566 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 11 |
Hb_001396_010 |
0.1320034013 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000297_030 |
0.132186567 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000866_140 |
0.1333173151 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 14 |
Hb_000029_170 |
0.1340175467 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 15 |
Hb_001195_220 |
0.1353358649 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_008725_050 |
0.1362108603 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_024758_020 |
0.1382064915 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 18 |
Hb_003428_090 |
0.1430944104 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 19 |
Hb_001369_470 |
0.1432494129 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 20 |
Hb_016734_020 |
0.1436816489 |
- |
- |
sugar transporter, putative [Ricinus communis] |