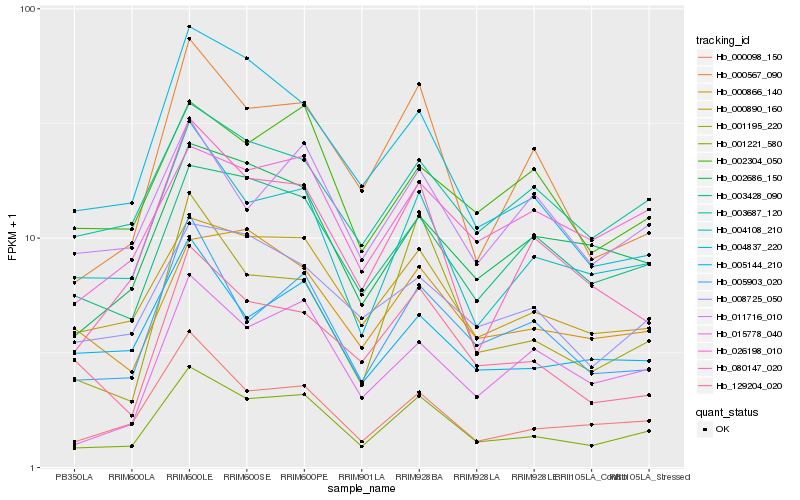
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_580 |
0.0 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 2 |
Hb_004108_210 |
0.0916987074 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 3 |
Hb_000098_150 |
0.0965867614 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 4 |
Hb_000890_160 |
0.1007509345 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 5 |
Hb_000567_090 |
0.1098748948 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_003428_090 |
0.1130777649 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 7 |
Hb_005903_020 |
0.1151819362 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 8 |
Hb_129204_020 |
0.118064363 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 9 |
Hb_080147_020 |
0.1192933776 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 10 |
Hb_002686_150 |
0.1195201342 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_001195_220 |
0.1200505078 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 12 |
Hb_003687_120 |
0.1208951502 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 13 |
Hb_015778_040 |
0.1234169577 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 14 |
Hb_005144_210 |
0.1235320943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000866_140 |
0.1260673327 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 16 |
Hb_008725_050 |
0.1268742006 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 17 |
Hb_002304_050 |
0.1302214567 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 18 |
Hb_026198_010 |
0.1308714403 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004837_220 |
0.131438898 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 20 |
Hb_011716_010 |
0.132391995 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |