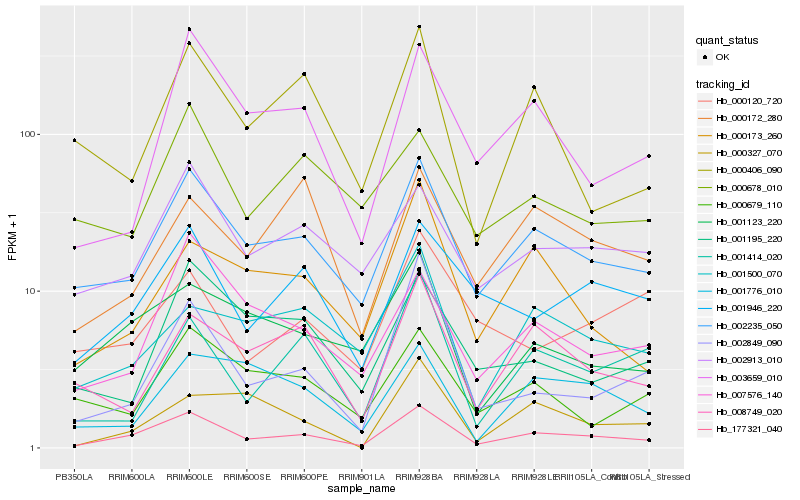
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177321_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_06328 [Jatropha curcas] |
| 2 |
Hb_002235_050 |
0.1231430092 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001946_220 |
0.1761953803 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 4 |
Hb_002913_010 |
0.1801077537 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003659_010 |
0.1847228456 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 6 |
Hb_001123_220 |
0.1875822882 |
- |
- |
- |
| 7 |
Hb_007576_140 |
0.1924000664 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 8 |
Hb_002849_090 |
0.1947444896 |
- |
- |
hypothetical protein POPTR_0002s16380g [Populus trichocarpa] |
| 9 |
Hb_008749_020 |
0.1964933882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000173_260 |
0.197335618 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 11 |
Hb_001414_020 |
0.1985457279 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001500_070 |
0.2031023017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000406_090 |
0.2070813288 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_000679_110 |
0.210095092 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 15 |
Hb_000172_280 |
0.2127701185 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 16 |
Hb_001195_220 |
0.2148489028 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_001776_010 |
0.2155143056 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 18 |
Hb_000678_010 |
0.2168182942 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000327_070 |
0.219923998 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000120_720 |
0.2209525521 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |