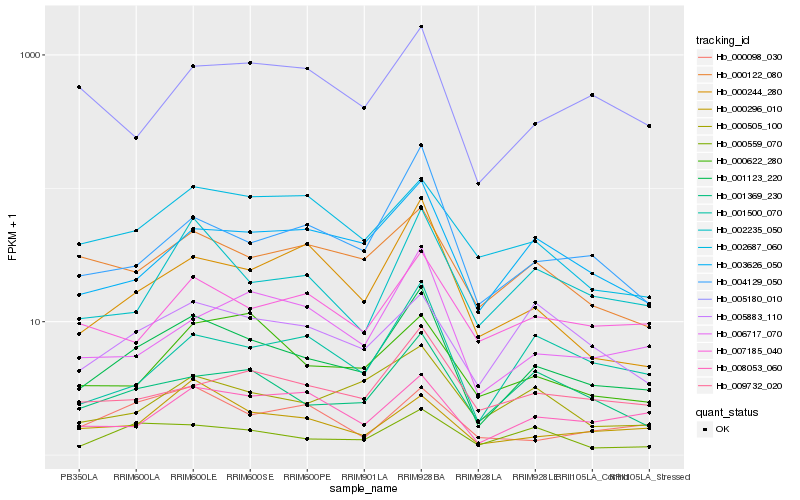
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_001369_230 |
0.1465591835 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 3 |
Hb_000559_070 |
0.1500949073 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 4 |
Hb_003626_050 |
0.1510223479 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 5 |
Hb_002235_050 |
0.1548280381 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004129_050 |
0.1549411817 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 7 |
Hb_009732_020 |
0.1611001625 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 8 |
Hb_001500_070 |
0.1617385344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008053_060 |
0.1618899072 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006717_070 |
0.1623945297 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 11 |
Hb_000505_100 |
0.1658080063 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100250663 [Vitis vinifera] |
| 12 |
Hb_000244_280 |
0.1664485343 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007185_040 |
0.1692804106 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 14 |
Hb_000122_080 |
0.1699029044 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 15 |
Hb_000296_010 |
0.1699381812 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 16 |
Hb_000622_280 |
0.1747358196 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005883_110 |
0.1756206953 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002687_060 |
0.1774166439 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 19 |
Hb_005180_010 |
0.179823922 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 20 |
Hb_000098_030 |
0.1798585359 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |