| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
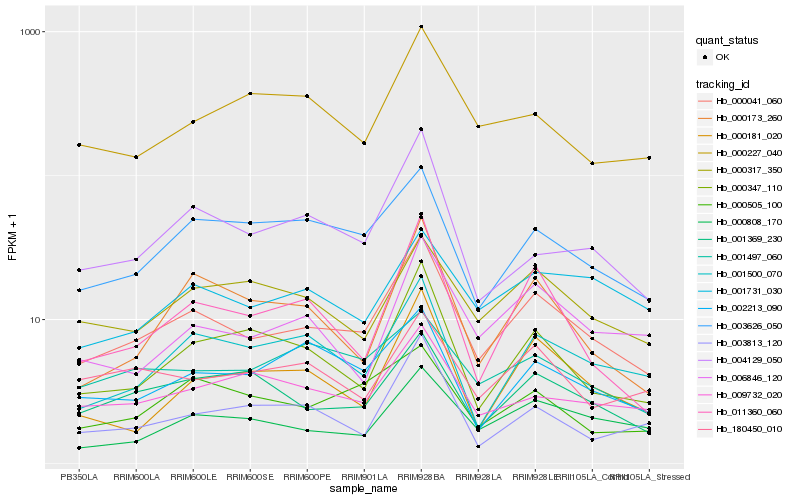
Hb_000041_060 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_000808_170 |
0.1276549756 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 3 |
Hb_001369_230 |
0.1329959467 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 4 |
Hb_001500_070 |
0.1334304869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003626_050 |
0.1344796001 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 6 |
Hb_000347_110 |
0.1361027916 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 7 |
Hb_004129_050 |
0.1376484565 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 8 |
Hb_006846_120 |
0.1390006485 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 9 |
Hb_000317_350 |
0.1414473425 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 10 |
Hb_011360_060 |
0.1437206346 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 11 |
Hb_000181_020 |
0.144108201 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000505_100 |
0.1442705654 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100250663 [Vitis vinifera] |
| 13 |
Hb_009732_020 |
0.1442765151 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_002213_090 |
0.1453409082 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 15 |
Hb_000173_260 |
0.1462072304 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 16 |
Hb_001731_030 |
0.147882616 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 17 |
Hb_180450_010 |
0.1505168318 |
- |
- |
FF domain-containing family protein [Populus trichocarpa] |
| 18 |
Hb_000227_040 |
0.1514052022 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 19 |
Hb_001497_060 |
0.1538839494 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_003813_120 |
0.1567396422 |
- |
- |
zinc finger protein, putative [Ricinus communis] |