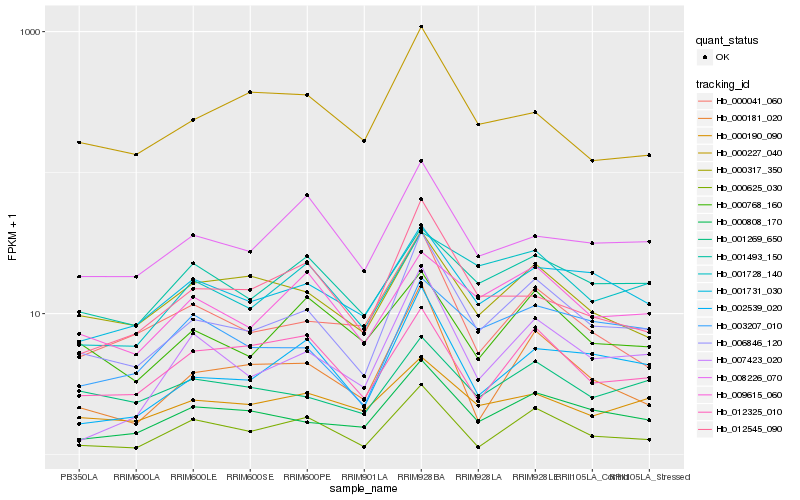
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006846_120 |
0.0 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 2 |
Hb_000808_170 |
0.1082461596 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 3 |
Hb_002539_020 |
0.125111568 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 4 |
Hb_001731_030 |
0.1319277597 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 5 |
Hb_000181_020 |
0.1329921949 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000625_030 |
0.1371923538 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 7 |
Hb_007423_020 |
0.1389667576 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 8 |
Hb_000041_060 |
0.1390006485 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_000317_350 |
0.1440878048 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 10 |
Hb_001269_650 |
0.1456357241 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_008226_070 |
0.1462192441 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 12 |
Hb_012545_090 |
0.1469068527 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 13 |
Hb_009615_060 |
0.1511533067 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 14 |
Hb_003207_010 |
0.152230893 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 15 |
Hb_000190_090 |
0.1527037436 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 16 |
Hb_000768_160 |
0.1529560786 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 17 |
Hb_001493_150 |
0.1542950788 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 18 |
Hb_012325_010 |
0.1558662736 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 19 |
Hb_000227_040 |
0.1573460053 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 20 |
Hb_001728_140 |
0.158128913 |
- |
- |
- |