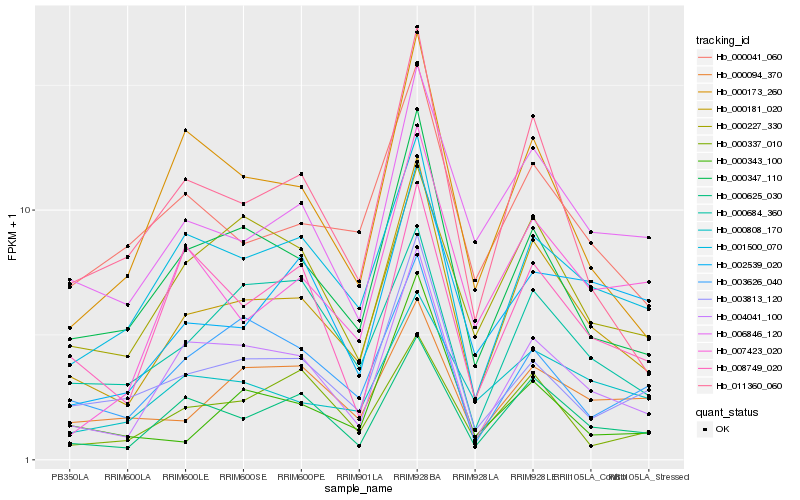
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_020 |
0.0 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_004041_100 |
0.1042843103 |
- |
- |
- |
| 3 |
Hb_000625_030 |
0.1111544713 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 4 |
Hb_000347_110 |
0.1119438343 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 5 |
Hb_011360_060 |
0.1215841099 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 6 |
Hb_006846_120 |
0.1329921949 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 7 |
Hb_001500_070 |
0.1351748997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000173_260 |
0.1405780246 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 9 |
Hb_000041_060 |
0.144108201 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_003813_120 |
0.1474386729 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000094_370 |
0.1552315365 |
- |
- |
- |
| 12 |
Hb_000808_170 |
0.1564454457 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 13 |
Hb_000684_360 |
0.1578730997 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 14 |
Hb_000227_330 |
0.1594466811 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 15 |
Hb_007423_020 |
0.1609322397 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 16 |
Hb_008749_020 |
0.162430198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002539_020 |
0.1634731851 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 18 |
Hb_000343_100 |
0.1648981042 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 19 |
Hb_003626_040 |
0.1654299388 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000337_010 |
0.1657849369 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |