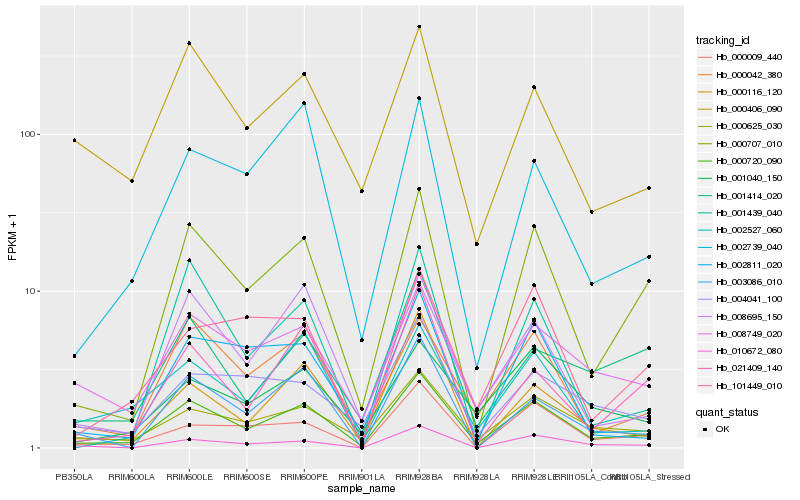
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000720_090 |
0.0 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 2 |
Hb_000707_010 |
0.1157868867 |
- |
- |
hypothetical protein JCGZ_25098 [Jatropha curcas] |
| 3 |
Hb_000625_030 |
0.1272698492 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 4 |
Hb_008749_020 |
0.1327572923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000042_380 |
0.1433928229 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 6 |
Hb_002527_060 |
0.1536398468 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 7 |
Hb_008695_150 |
0.1611160408 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000009_440 |
0.1629139626 |
- |
- |
PREDICTED: uncharacterized protein LOC105639617 [Jatropha curcas] |
| 9 |
Hb_002739_040 |
0.1630487654 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 10 |
Hb_021409_140 |
0.1642028498 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 11 |
Hb_001040_150 |
0.164816325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000406_090 |
0.1655948947 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_010672_080 |
0.1663051922 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 14 |
Hb_003086_010 |
0.1666756584 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 15 |
Hb_004041_100 |
0.168013456 |
- |
- |
- |
| 16 |
Hb_001414_020 |
0.1695897536 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002811_020 |
0.1706983435 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 18 |
Hb_001439_040 |
0.1713748116 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_101449_010 |
0.1722807126 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 20 |
Hb_000116_120 |
0.172925736 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |