| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
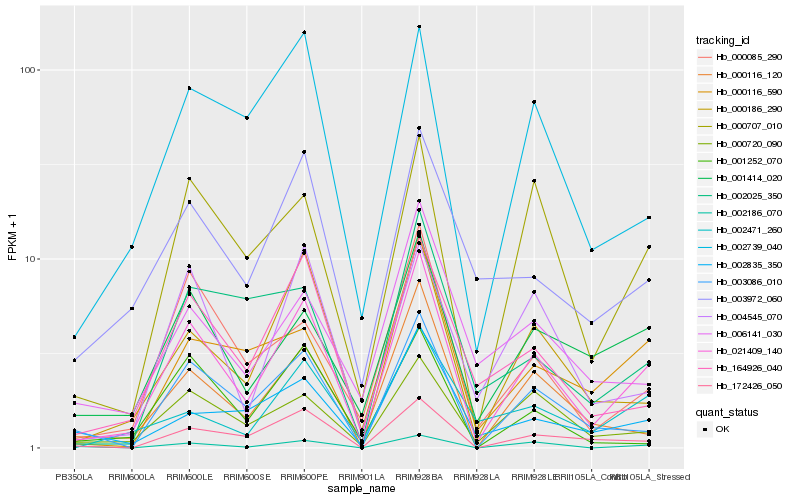
Hb_021409_140 |
0.0 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 2 |
Hb_000186_290 |
0.1577014419 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000116_120 |
0.1634781409 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 4 |
Hb_000707_010 |
0.1639660771 |
- |
- |
hypothetical protein JCGZ_25098 [Jatropha curcas] |
| 5 |
Hb_000720_090 |
0.1642028498 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 6 |
Hb_003086_010 |
0.1703320781 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 7 |
Hb_172426_050 |
0.1719844319 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 8 |
Hb_001414_020 |
0.1743095209 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_164926_040 |
0.1751332414 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 10 |
Hb_000085_290 |
0.1792138395 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 11 |
Hb_002739_040 |
0.1851908355 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 12 |
Hb_002186_070 |
0.186743967 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_000116_590 |
0.1902751611 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 14 |
Hb_004545_070 |
0.1961211706 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_003972_060 |
0.1970977003 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 16 |
Hb_002835_350 |
0.200385881 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 17 |
Hb_002471_260 |
0.2007206054 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 18 |
Hb_001252_070 |
0.20186789 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 19 |
Hb_006141_030 |
0.2077356878 |
- |
- |
- |
| 20 |
Hb_002025_350 |
0.2083642444 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |