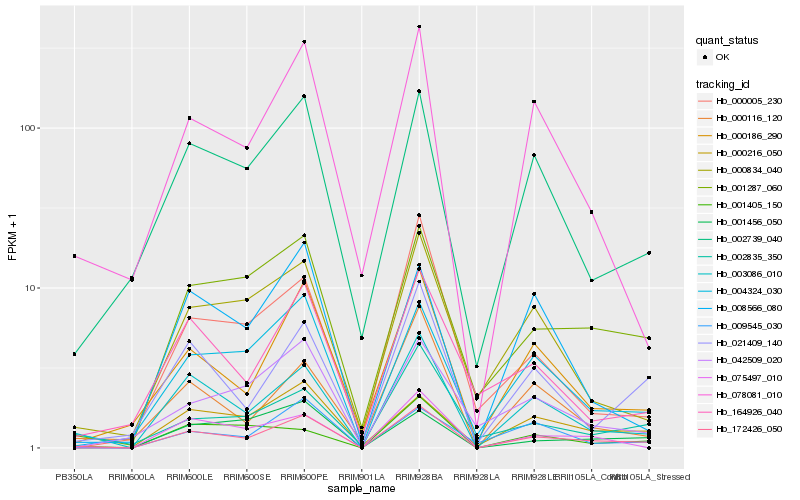
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172426_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 2 |
Hb_003086_010 |
0.1145586628 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 3 |
Hb_000186_290 |
0.1578164187 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001287_060 |
0.1583622582 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 5 |
Hb_004324_030 |
0.1622562118 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 6 |
Hb_021409_140 |
0.1719844319 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 7 |
Hb_002739_040 |
0.17198679 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 8 |
Hb_078081_010 |
0.1740996294 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 9 |
Hb_000216_050 |
0.1809468596 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 10 |
Hb_000116_120 |
0.1843586123 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 11 |
Hb_075497_010 |
0.1879463961 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 12 |
Hb_000834_040 |
0.1916350979 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 13 |
Hb_009545_030 |
0.1933867629 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_164926_040 |
0.1964764313 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 15 |
Hb_002835_350 |
0.1979859549 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 16 |
Hb_001405_150 |
0.1999687691 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 17 |
Hb_001456_050 |
0.2023411657 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_042509_020 |
0.2044700749 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 19 |
Hb_000005_230 |
0.209705087 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 20 |
Hb_008566_080 |
0.2098040409 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |