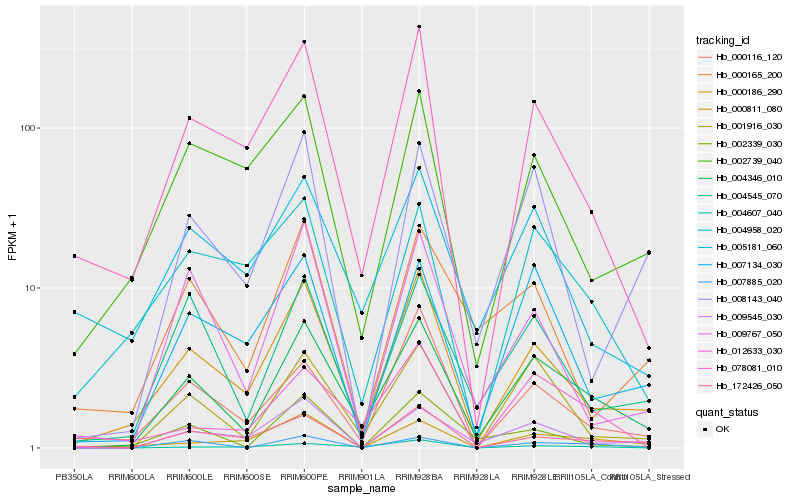
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004346_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000186_290 |
0.1516643348 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001916_030 |
0.1564713498 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 4 |
Hb_004545_070 |
0.1756113852 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_007885_020 |
0.1868005298 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 6 |
Hb_009545_030 |
0.1941974474 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_078081_010 |
0.1959113861 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 8 |
Hb_007134_030 |
0.1971798756 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 9 |
Hb_008143_040 |
0.2083279662 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 10 |
Hb_009767_050 |
0.2104402957 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005181_060 |
0.2171952315 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 12 |
Hb_000811_080 |
0.2196660674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004607_040 |
0.2198088557 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 14 |
Hb_012633_030 |
0.2222453843 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 15 |
Hb_000116_120 |
0.2227384544 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 16 |
Hb_172426_050 |
0.2242004083 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 17 |
Hb_002739_040 |
0.2243920682 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 18 |
Hb_002339_030 |
0.225110629 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000165_200 |
0.2252590604 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_004958_020 |
0.2262583908 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |