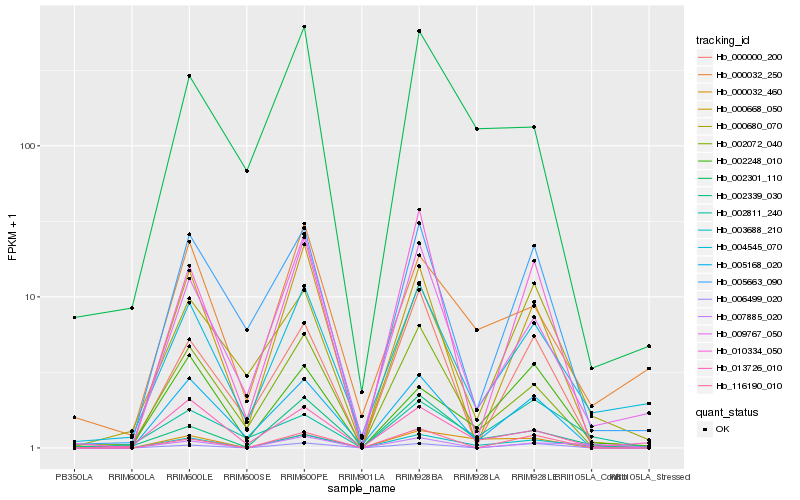
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_210 |
0.0 |
- |
- |
replication factor A 1, rfa1, putative [Ricinus communis] |
| 2 |
Hb_004545_070 |
0.1746599512 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000032_460 |
0.187003857 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 4 |
Hb_002339_030 |
0.1907136689 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002072_040 |
0.1984158543 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 6 |
Hb_005168_020 |
0.1998491943 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 7 |
Hb_000668_050 |
0.2068973134 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 8 |
Hb_009767_050 |
0.2079666166 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007885_020 |
0.2127821303 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 10 |
Hb_002248_010 |
0.2147600148 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 11 |
Hb_002811_240 |
0.2193740832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000032_250 |
0.2194749069 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 13 |
Hb_000680_070 |
0.2201986768 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_116190_010 |
0.2212850614 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 15 |
Hb_010334_050 |
0.2213027003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005663_090 |
0.2234303849 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 17 |
Hb_013726_010 |
0.225537384 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_006499_020 |
0.2282440745 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 19 |
Hb_000000_200 |
0.2317501313 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 20 |
Hb_002301_110 |
0.233500132 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |