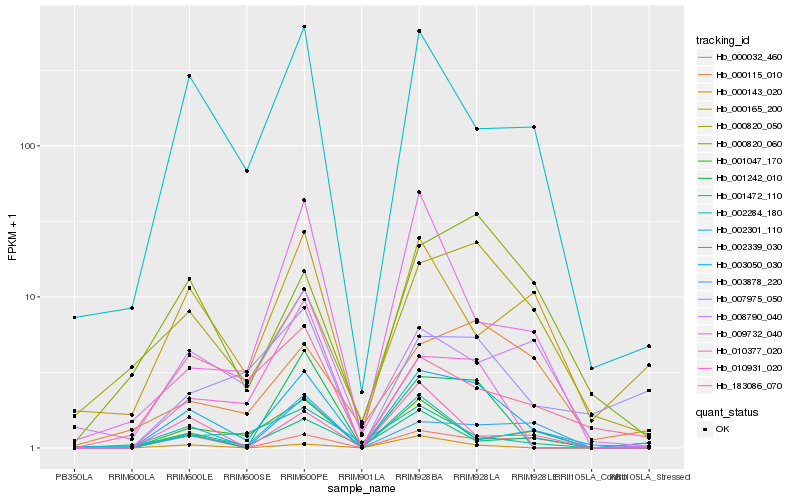
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_030 |
0.0 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 2 |
Hb_002301_110 |
0.2202412605 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 3 |
Hb_000032_460 |
0.2446131548 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 4 |
Hb_003878_220 |
0.2467211426 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 5 |
Hb_001242_010 |
0.2519959996 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 6 |
Hb_001472_110 |
0.254520342 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 7 |
Hb_007975_050 |
0.2595297785 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 8 |
Hb_000820_050 |
0.2661668076 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 9 |
Hb_000820_060 |
0.2686989477 |
- |
- |
- |
| 10 |
Hb_000165_200 |
0.270523057 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_010377_020 |
0.2717801171 |
- |
- |
- |
| 12 |
Hb_000115_010 |
0.2744585204 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001047_170 |
0.2756223754 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 14 |
Hb_002339_030 |
0.2823208112 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_010931_020 |
0.2838846768 |
- |
- |
Kunitz-type protease inhibitor KPI-F4 [Populus trichocarpa x Populus deltoides] |
| 16 |
Hb_008790_040 |
0.2870915565 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 17 |
Hb_009732_040 |
0.2943966849 |
- |
- |
hypothetical protein POPTR_0006s04880g [Populus trichocarpa] |
| 18 |
Hb_002284_180 |
0.2958159949 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 19 |
Hb_183086_070 |
0.2961852675 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 20 |
Hb_000143_020 |
0.2981592316 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |