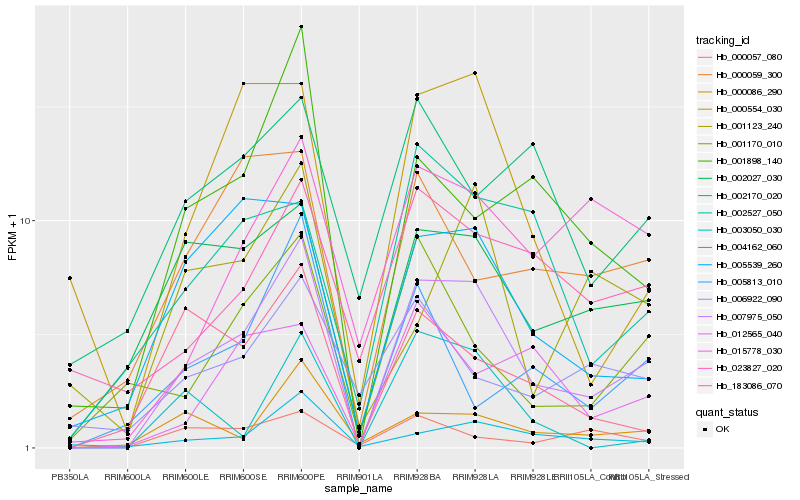
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007975_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 2 |
Hb_002027_030 |
0.2131706681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004162_060 |
0.2235696904 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000086_290 |
0.2319665678 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 5 |
Hb_005539_260 |
0.2376025011 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 6 |
Hb_000554_030 |
0.2379150128 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_015778_030 |
0.2400080337 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 8 |
Hb_023827_020 |
0.2415886832 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 9 |
Hb_001170_010 |
0.2464883514 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001898_140 |
0.2465929188 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 11 |
Hb_183086_070 |
0.2532072314 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 12 |
Hb_002527_050 |
0.2589891442 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 13 |
Hb_003050_030 |
0.2595297785 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 14 |
Hb_012565_040 |
0.2603366973 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 15 |
Hb_000059_300 |
0.264106992 |
- |
- |
caspase, putative [Ricinus communis] |
| 16 |
Hb_000057_080 |
0.2655959831 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 17 |
Hb_005813_010 |
0.2657437519 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_006922_090 |
0.2668236696 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 19 |
Hb_001123_240 |
0.2679430607 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 20 |
Hb_002170_020 |
0.269163855 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |