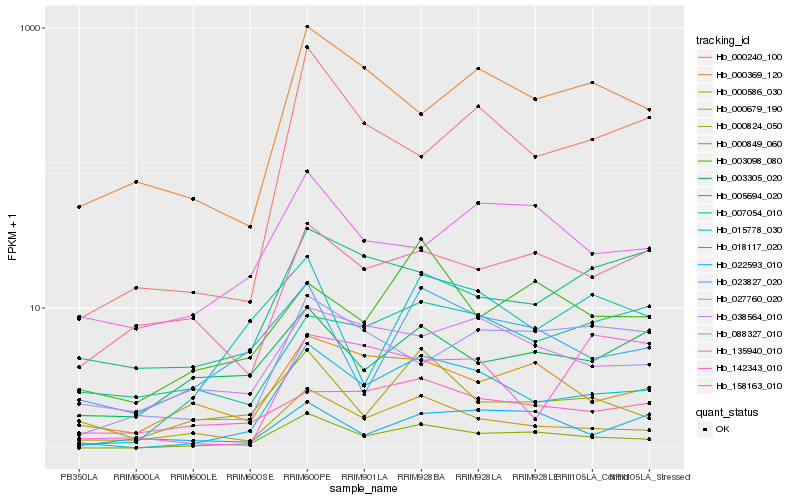
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018117_020 |
0.0 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_000849_060 |
0.1336801903 |
- |
- |
putative sodium-dependent transporter yocS [Morus notabilis] |
| 3 |
Hb_000679_190 |
0.1944657655 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000240_100 |
0.2127181817 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 5 |
Hb_015778_030 |
0.2149578659 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 6 |
Hb_038564_010 |
0.2150476643 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 7 |
Hb_005694_020 |
0.2241047221 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 8 |
Hb_000369_120 |
0.2246729085 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 9 |
Hb_007054_010 |
0.2249850585 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 10 |
Hb_000586_030 |
0.2305516679 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 11 |
Hb_027760_020 |
0.2324632405 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |
| 12 |
Hb_158163_010 |
0.2343618611 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_142343_010 |
0.2350357474 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_023827_020 |
0.2352107348 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 15 |
Hb_022593_010 |
0.2421708553 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003305_020 |
0.2427015174 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 17 |
Hb_000824_050 |
0.2442839921 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_003098_080 |
0.2447475743 |
- |
- |
- |
| 19 |
Hb_135940_010 |
0.246126824 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 20 |
Hb_088327_010 |
0.2483452714 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |