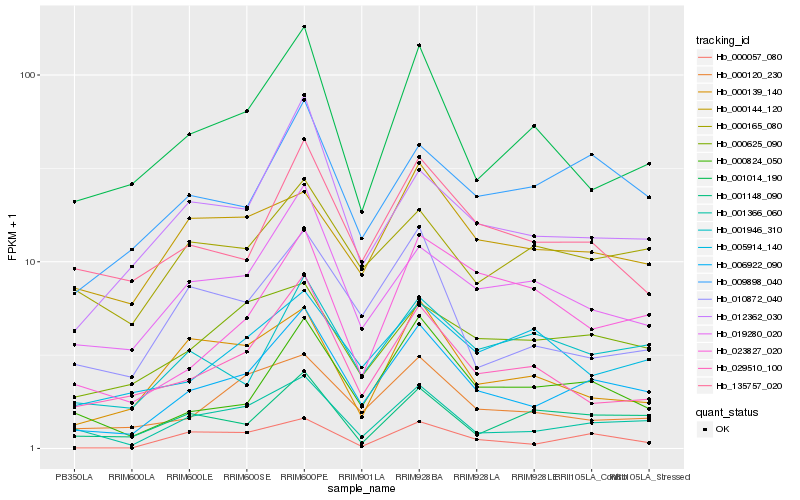
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006922_090 |
0.0 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 2 |
Hb_001366_060 |
0.1236696852 |
- |
- |
- |
| 3 |
Hb_000824_050 |
0.1354245641 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_012362_030 |
0.142929619 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 5 |
Hb_000120_230 |
0.1456259539 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 6 |
Hb_029510_100 |
0.1516753957 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 7 |
Hb_019280_020 |
0.1517450575 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001014_190 |
0.1527852344 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 9 |
Hb_000057_080 |
0.1549803481 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 10 |
Hb_009898_040 |
0.1558722692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_135757_020 |
0.157995299 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 12 |
Hb_001946_310 |
0.1611458383 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 13 |
Hb_010872_040 |
0.1632504548 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 14 |
Hb_000625_090 |
0.1692611768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000139_140 |
0.1749166953 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 16 |
Hb_005914_140 |
0.177455167 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 17 |
Hb_023827_020 |
0.1784951682 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 18 |
Hb_000165_080 |
0.1794141256 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001148_090 |
0.1805112865 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 20 |
Hb_000144_120 |
0.1815619922 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |