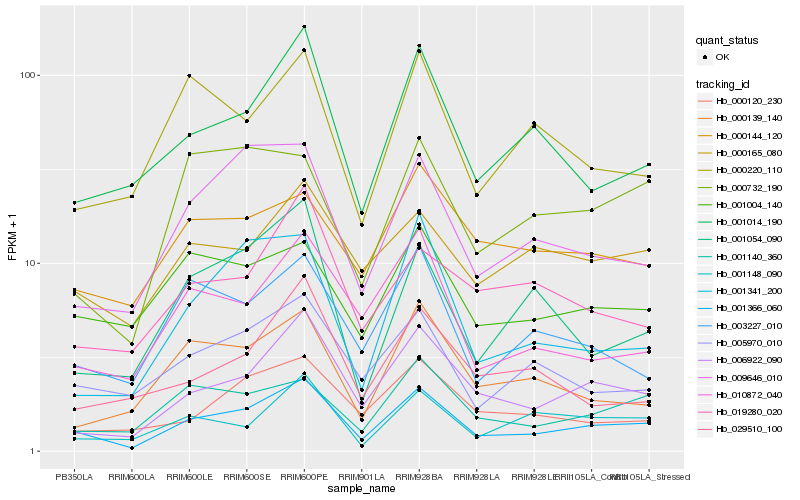
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_006922_090 |
0.1236696852 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 3 |
Hb_001014_190 |
0.1474087929 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 4 |
Hb_009646_010 |
0.1494026891 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 5 |
Hb_005970_010 |
0.1537272254 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010872_040 |
0.1566628458 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 7 |
Hb_001140_360 |
0.1584284962 |
- |
- |
- |
| 8 |
Hb_000120_230 |
0.1621704666 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 9 |
Hb_001341_200 |
0.1630398255 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 10 |
Hb_003227_010 |
0.1642003111 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 11 |
Hb_000165_080 |
0.1672183826 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001054_090 |
0.1676432476 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 13 |
Hb_001004_140 |
0.1680714861 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 14 |
Hb_000139_140 |
0.1706161081 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 15 |
Hb_001148_090 |
0.1709874338 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 16 |
Hb_000732_190 |
0.1715154121 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 17 |
Hb_000220_110 |
0.1727842634 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 18 |
Hb_000144_120 |
0.1741588488 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_019280_020 |
0.1741781059 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 20 |
Hb_029510_100 |
0.1806051909 |
- |
- |
scab resistance family protein [Populus trichocarpa] |