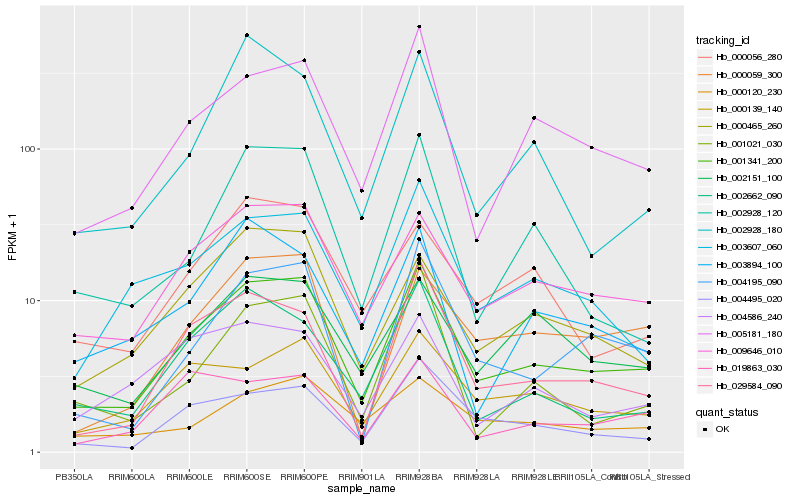
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001341_200 |
0.0 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 2 |
Hb_009646_010 |
0.1111194412 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 3 |
Hb_005181_180 |
0.1113877287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004495_020 |
0.125360064 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 5 |
Hb_029584_090 |
0.1279642766 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 6 |
Hb_004195_090 |
0.1315617382 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 7 |
Hb_000139_140 |
0.1383991987 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 8 |
Hb_003607_060 |
0.1386680719 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 9 |
Hb_000465_260 |
0.1398508274 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 10 |
Hb_002662_090 |
0.1447578999 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 11 |
Hb_002151_100 |
0.1465848576 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 12 |
Hb_000059_300 |
0.1515725599 |
- |
- |
caspase, putative [Ricinus communis] |
| 13 |
Hb_003894_100 |
0.1519287147 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 14 |
Hb_002928_120 |
0.1542678292 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 15 |
Hb_001021_030 |
0.1582744162 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_000120_230 |
0.158744698 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 17 |
Hb_000056_280 |
0.1609677597 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 18 |
Hb_019863_030 |
0.1613337574 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 19 |
Hb_004586_240 |
0.1614451592 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 20 |
Hb_002928_180 |
0.1621665293 |
- |
- |
catalase CAT1 [Manihot esculenta] |