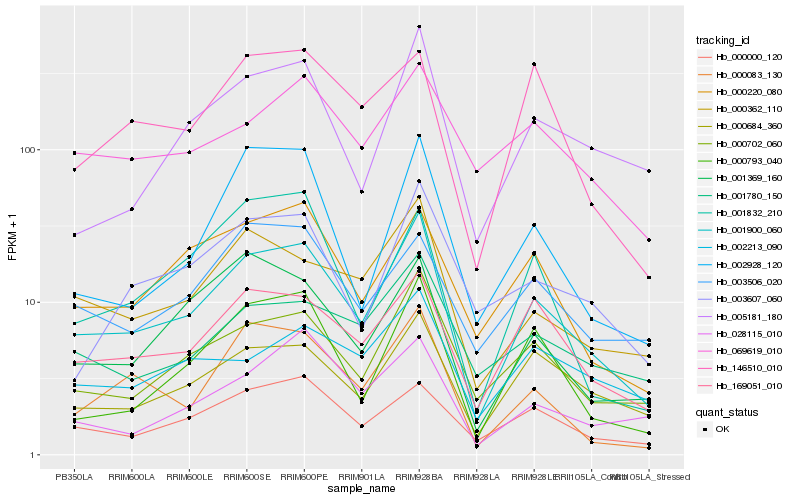
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001900_060 |
0.0 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 2 |
Hb_000702_060 |
0.1195892307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000684_360 |
0.1307557254 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 4 |
Hb_169051_010 |
0.1327006547 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 5 |
Hb_000362_110 |
0.1360516959 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 6 |
Hb_000793_040 |
0.1365939418 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 7 |
Hb_000083_130 |
0.1392264696 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 8 |
Hb_000000_120 |
0.1411012034 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 9 |
Hb_005181_180 |
0.1497506396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000220_080 |
0.1506133635 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_001369_160 |
0.1529521796 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 12 |
Hb_002928_120 |
0.1542295962 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 13 |
Hb_003607_060 |
0.1569963273 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 14 |
Hb_001780_150 |
0.1578707312 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 15 |
Hb_146510_010 |
0.1648652244 |
- |
- |
cinnamyl alcohol dehydrogenase [Hevea brasiliensis] |
| 16 |
Hb_028115_010 |
0.1650300602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003506_020 |
0.1660366621 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 18 |
Hb_069619_010 |
0.1683142129 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 19 |
Hb_001832_210 |
0.1696214438 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 20 |
Hb_002213_090 |
0.1701359807 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |