| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
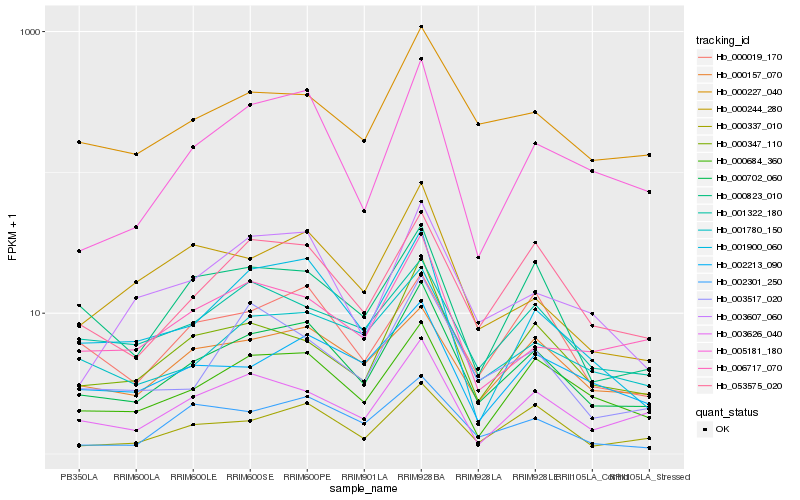
| 1 |
Hb_000702_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000684_360 |
0.1117847825 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 3 |
Hb_053575_020 |
0.1122822277 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 4 |
Hb_000337_010 |
0.1156411721 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000347_110 |
0.1180395838 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 6 |
Hb_001780_150 |
0.1188615572 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 7 |
Hb_005181_180 |
0.1191439788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001900_060 |
0.1195892307 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 9 |
Hb_003626_040 |
0.1227809992 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000227_040 |
0.1230189707 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 11 |
Hb_000157_070 |
0.1291786274 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 12 |
Hb_002301_250 |
0.13144078 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a [Jatropha curcas] |
| 13 |
Hb_000823_010 |
0.13209879 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 14 |
Hb_002213_090 |
0.1362784911 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 15 |
Hb_006717_070 |
0.1367850661 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 16 |
Hb_003517_020 |
0.137917083 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 17 |
Hb_003607_060 |
0.1382282513 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_000244_280 |
0.1385883335 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001322_180 |
0.1388341698 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 20 |
Hb_000019_170 |
0.1394436202 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |