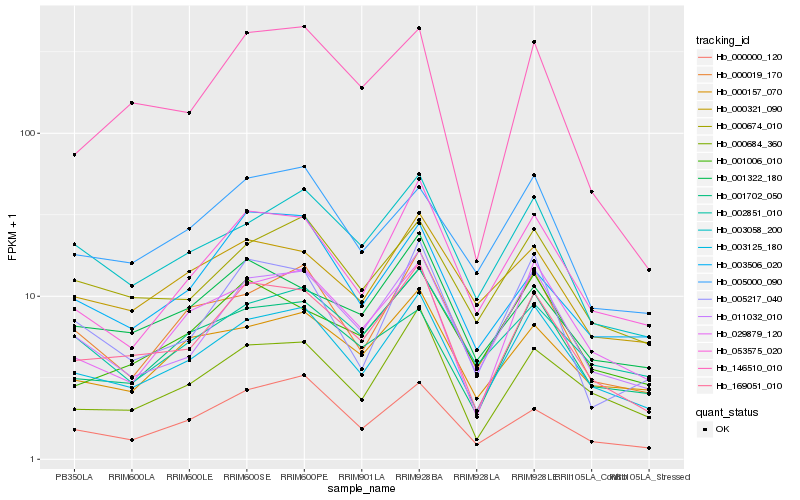
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169051_010 |
0.0 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 2 |
Hb_003125_180 |
0.0776969252 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 3 |
Hb_011032_010 |
0.0893835827 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_000684_360 |
0.097931557 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 5 |
Hb_146510_010 |
0.0990394596 |
- |
- |
cinnamyl alcohol dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_001322_180 |
0.1038983203 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 7 |
Hb_053575_020 |
0.1127470009 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 8 |
Hb_000000_120 |
0.1163069238 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 9 |
Hb_000674_010 |
0.1169866975 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003058_200 |
0.1177888157 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 11 |
Hb_001006_010 |
0.1182111287 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 12 |
Hb_029879_120 |
0.1184888004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000157_070 |
0.1241073125 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 14 |
Hb_005217_040 |
0.1256820751 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000321_090 |
0.1282590251 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_000019_170 |
0.1298036274 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 17 |
Hb_001702_050 |
0.1302082766 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003506_020 |
0.130223539 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 19 |
Hb_005000_090 |
0.1305620654 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 20 |
Hb_002851_010 |
0.1312949623 |
- |
- |
sugar transporter, putative [Ricinus communis] |