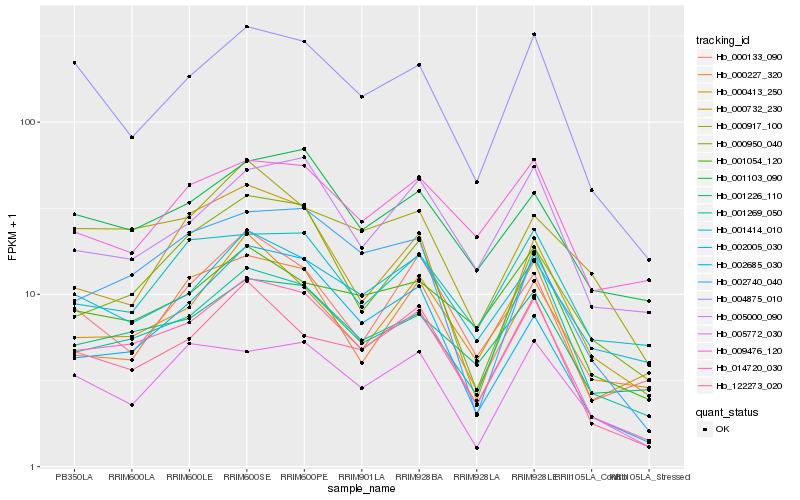
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014720_030 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 2 |
Hb_004875_010 |
0.0970066466 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 3 |
Hb_001226_110 |
0.0987260479 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 4 |
Hb_001269_050 |
0.1012411726 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 5 |
Hb_122273_020 |
0.102378686 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 6 |
Hb_001103_090 |
0.1073025395 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 7 |
Hb_005000_090 |
0.1120499851 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 8 |
Hb_002005_030 |
0.1142876388 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 9 |
Hb_002740_040 |
0.1152623081 |
- |
- |
hypothetical protein POPTR_0005s00350g [Populus trichocarpa] |
| 10 |
Hb_000950_040 |
0.116523869 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 11 |
Hb_001054_120 |
0.118612348 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 12 |
Hb_002685_030 |
0.1188611263 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 13 |
Hb_000917_100 |
0.1190512167 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 14 |
Hb_000732_230 |
0.1228303295 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000133_090 |
0.1229561142 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 16 |
Hb_000413_250 |
0.1241994487 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 17 |
Hb_000227_320 |
0.1249279077 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001414_010 |
0.1252991816 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 19 |
Hb_009476_120 |
0.1278366221 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 20 |
Hb_005772_030 |
0.128108755 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |