| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
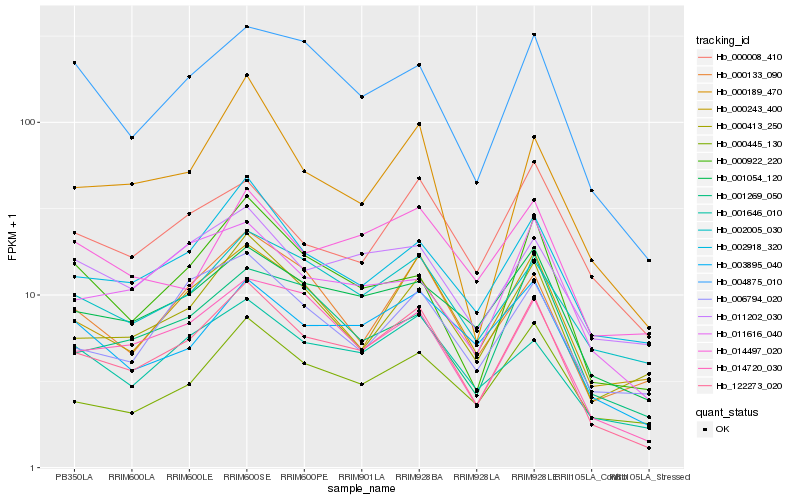
Hb_122273_020 |
0.0 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 2 |
Hb_001054_120 |
0.1018174415 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 3 |
Hb_014720_030 |
0.102378686 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 4 |
Hb_000243_400 |
0.1086349929 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 5 |
Hb_002918_320 |
0.1092417225 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004875_010 |
0.1119160503 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 7 |
Hb_000413_250 |
0.1124753882 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 8 |
Hb_003895_040 |
0.1150358696 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |
| 9 |
Hb_001646_010 |
0.1151383122 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 10 |
Hb_000922_220 |
0.1166352788 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 11 |
Hb_002005_030 |
0.1187248394 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 12 |
Hb_000133_090 |
0.1219585193 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 13 |
Hb_000445_130 |
0.1232889478 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 14 |
Hb_000189_470 |
0.1233874947 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 15 |
Hb_006794_020 |
0.1251693316 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 16 |
Hb_011616_040 |
0.1263928992 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 17 |
Hb_001269_050 |
0.1270242208 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 18 |
Hb_011202_030 |
0.1276153796 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 19 |
Hb_000008_410 |
0.1294071065 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_014497_020 |
0.1303670333 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |