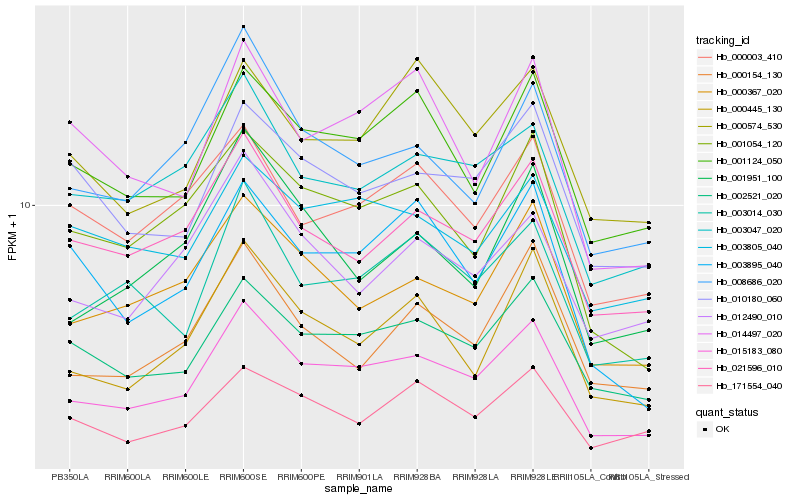
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015183_080 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_002521_020 |
0.0818469896 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_003014_030 |
0.0859519974 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |
| 4 |
Hb_000445_130 |
0.0868955415 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 5 |
Hb_001054_120 |
0.0871647586 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 6 |
Hb_000367_020 |
0.0882789328 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 7 |
Hb_001124_050 |
0.0882836698 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 8 |
Hb_021596_010 |
0.0912021016 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 9 |
Hb_003047_020 |
0.0936662926 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_008686_020 |
0.0952735177 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003895_040 |
0.1005125791 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |
| 12 |
Hb_014497_020 |
0.1014547495 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 13 |
Hb_000003_410 |
0.1022071067 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 14 |
Hb_000154_130 |
0.1032293884 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 15 |
Hb_010180_060 |
0.1037220944 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 16 |
Hb_171554_040 |
0.1037507407 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 17 |
Hb_012490_010 |
0.105652997 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001951_100 |
0.1062307706 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 19 |
Hb_000574_530 |
0.1069271607 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 20 |
Hb_003805_040 |
0.10707228 |
- |
- |
Sacsin [Glycine soja] |