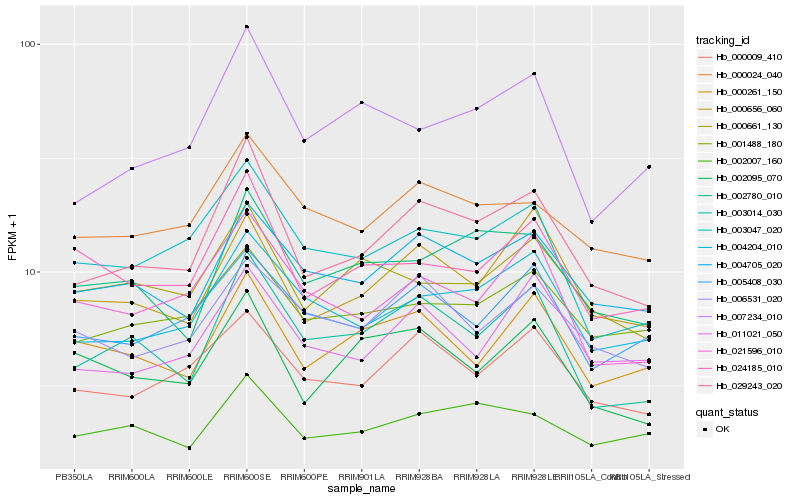
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029243_020 |
0.0 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 2 |
Hb_003014_030 |
0.086019226 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |
| 3 |
Hb_004705_020 |
0.0950794273 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 4 |
Hb_000009_410 |
0.0952877986 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 5 |
Hb_004204_010 |
0.0955386378 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 6 |
Hb_000656_060 |
0.0969411791 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_003047_020 |
0.1006054192 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_002780_010 |
0.1024597655 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 9 |
Hb_000024_040 |
0.1036127994 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 10 |
Hb_011021_050 |
0.104510047 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007234_010 |
0.1057565444 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 12 |
Hb_021596_010 |
0.1065534353 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 13 |
Hb_024185_010 |
0.1072679734 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002007_160 |
0.1116486763 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 15 |
Hb_001488_180 |
0.111675174 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000661_130 |
0.1118622549 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 17 |
Hb_005408_030 |
0.1122141883 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 18 |
Hb_002095_070 |
0.1132950257 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 19 |
Hb_000261_150 |
0.1138081767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006531_020 |
0.1153880228 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |