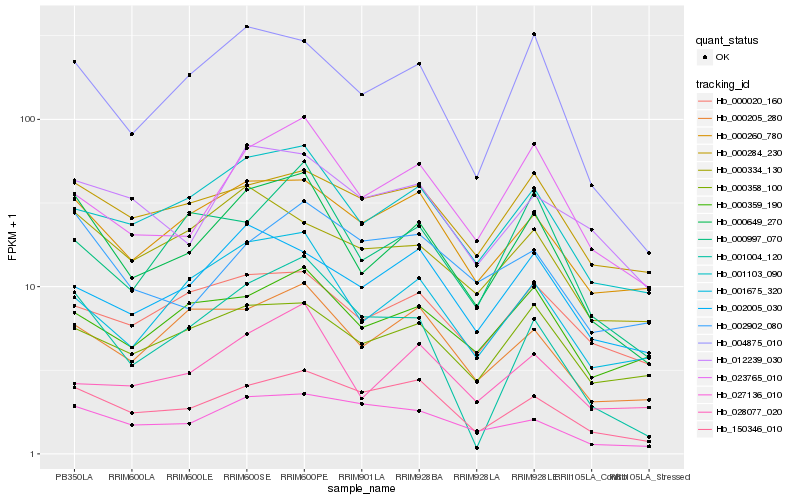
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_270 |
0.0 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_004875_010 |
0.114177687 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 3 |
Hb_001103_090 |
0.1220315337 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 4 |
Hb_001675_320 |
0.1296672533 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 5 |
Hb_000260_780 |
0.1305606071 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |
| 6 |
Hb_150346_010 |
0.1322776707 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 7 |
Hb_023765_010 |
0.1353588485 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 8 |
Hb_000205_280 |
0.1392630848 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_001004_120 |
0.1485668957 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000358_100 |
0.1513190171 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 11 |
Hb_000334_130 |
0.1518998502 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 12 |
Hb_027136_010 |
0.1532593121 |
- |
- |
hypothetical protein L484_010426 [Morus notabilis] |
| 13 |
Hb_012239_030 |
0.1532657696 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000997_070 |
0.1535165396 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 15 |
Hb_000359_190 |
0.1545989841 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 16 |
Hb_028077_020 |
0.1568429465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000020_160 |
0.1582140746 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 18 |
Hb_000284_230 |
0.1583408835 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 19 |
Hb_002005_030 |
0.1590166171 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_002902_080 |
0.1598437079 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |