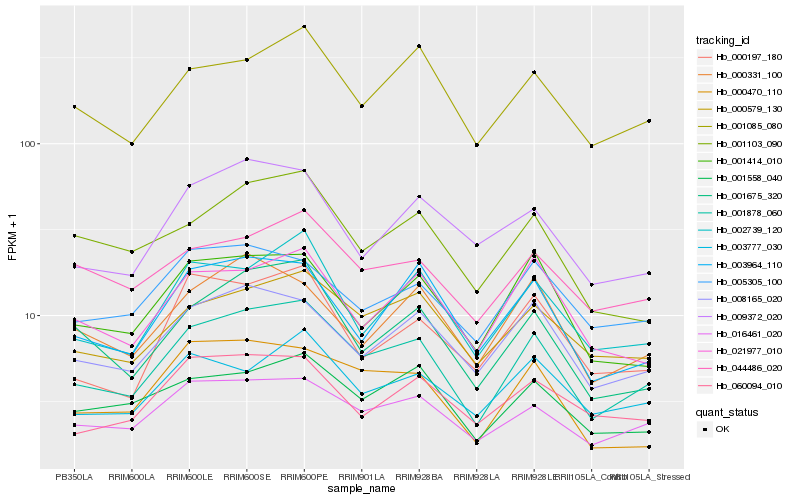
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001878_060 |
0.0 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0007s15110g [Populus trichocarpa] |
| 2 |
Hb_016461_020 |
0.0872623789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000579_130 |
0.0935086217 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_002739_120 |
0.0943908324 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 5 |
Hb_008165_020 |
0.0951391002 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001675_320 |
0.0954093289 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 7 |
Hb_001085_080 |
0.0988818914 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 8 |
Hb_001558_040 |
0.1007453937 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 9 |
Hb_000470_110 |
0.1042012019 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 10 |
Hb_003964_110 |
0.1049674239 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001414_010 |
0.1060842886 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 12 |
Hb_021977_010 |
0.1079362459 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 13 |
Hb_044486_020 |
0.1096305753 |
- |
- |
CASTOR protein [Glycine max] |
| 14 |
Hb_003777_030 |
0.1100903519 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 15 |
Hb_000331_100 |
0.1104273161 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000197_180 |
0.1120385695 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 17 |
Hb_001103_090 |
0.1121250683 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 18 |
Hb_005305_100 |
0.1125685456 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_009372_020 |
0.1127390641 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 20 |
Hb_060094_010 |
0.1128742159 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |