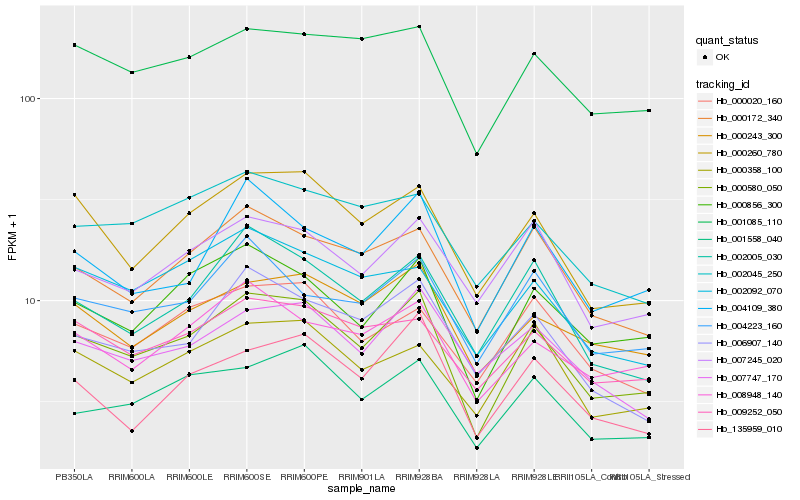
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000580_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000856_300 |
0.0755194034 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 3 |
Hb_000260_780 |
0.0834355788 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |
| 4 |
Hb_001558_040 |
0.0855772823 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 5 |
Hb_000243_300 |
0.0904128214 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_000172_340 |
0.0919493915 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 7 |
Hb_001085_110 |
0.094031946 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 8 |
Hb_008948_140 |
0.0948390888 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 9 |
Hb_002005_030 |
0.0957268764 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 10 |
Hb_004223_160 |
0.0970244523 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004109_380 |
0.0983424081 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 12 |
Hb_000020_160 |
0.0984338744 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 13 |
Hb_000358_100 |
0.0991988086 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 14 |
Hb_009252_050 |
0.0993173088 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_135959_010 |
0.1004656894 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 16 |
Hb_002045_250 |
0.1008038099 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 17 |
Hb_002092_070 |
0.100989683 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 18 |
Hb_007245_020 |
0.1012776728 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 19 |
Hb_006907_140 |
0.1014315524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007747_170 |
0.1024220022 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |