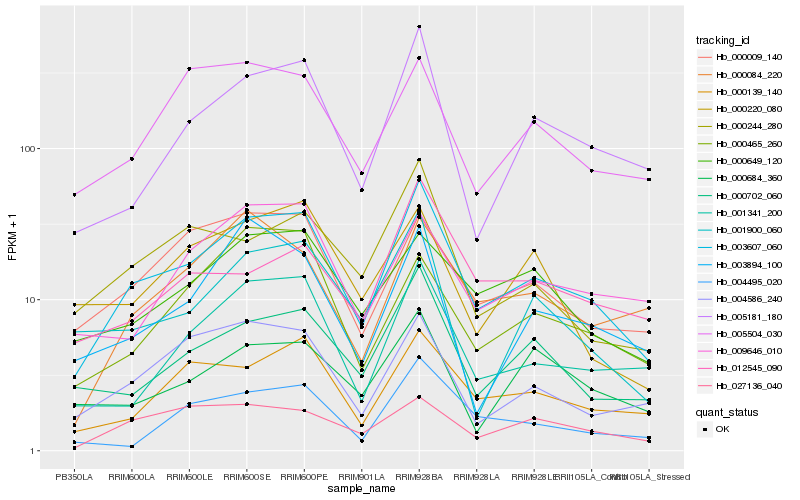
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003607_060 |
0.0 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 2 |
Hb_005181_180 |
0.1323161807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000702_060 |
0.1382282513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001341_200 |
0.1386680719 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 5 |
Hb_000139_140 |
0.1427894274 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 6 |
Hb_000009_140 |
0.1514327485 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 7 |
Hb_004586_240 |
0.1531998247 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 8 |
Hb_001900_060 |
0.1569963273 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 9 |
Hb_000244_280 |
0.1573362767 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000649_120 |
0.158893012 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_004495_020 |
0.1618404204 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 12 |
Hb_000084_220 |
0.1650666095 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 13 |
Hb_009646_010 |
0.1664679088 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 14 |
Hb_027136_040 |
0.1669732137 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 15 |
Hb_005504_030 |
0.1674885911 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 16 |
Hb_003894_100 |
0.168271581 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 17 |
Hb_000684_360 |
0.1714544626 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 18 |
Hb_000465_260 |
0.1723504576 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 19 |
Hb_012545_090 |
0.1736177839 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 20 |
Hb_000220_080 |
0.1740335985 |
- |
- |
kinase, putative [Ricinus communis] |