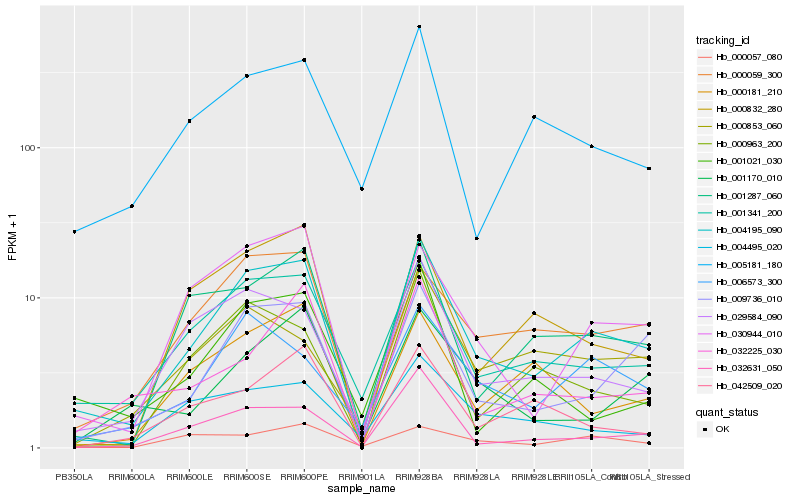
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004195_090 |
0.0 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 2 |
Hb_001341_200 |
0.1315617382 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 3 |
Hb_009736_010 |
0.1524470984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_029584_090 |
0.1604021997 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 5 |
Hb_030944_010 |
0.1615246479 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 6 |
Hb_000059_300 |
0.1638286493 |
- |
- |
caspase, putative [Ricinus communis] |
| 7 |
Hb_032631_050 |
0.1710687684 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_001287_060 |
0.1717389881 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 9 |
Hb_006573_300 |
0.1743258046 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_004495_020 |
0.1790710454 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_005181_180 |
0.1871700509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000832_280 |
0.187340175 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 13 |
Hb_000963_200 |
0.18860011 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 14 |
Hb_000181_210 |
0.1929069198 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 15 |
Hb_001170_010 |
0.1958336601 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000057_080 |
0.1979295283 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 17 |
Hb_000853_060 |
0.199053594 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_032225_030 |
0.1999148698 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 19 |
Hb_001021_030 |
0.2000788712 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 20 |
Hb_042509_020 |
0.2003004854 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |