| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
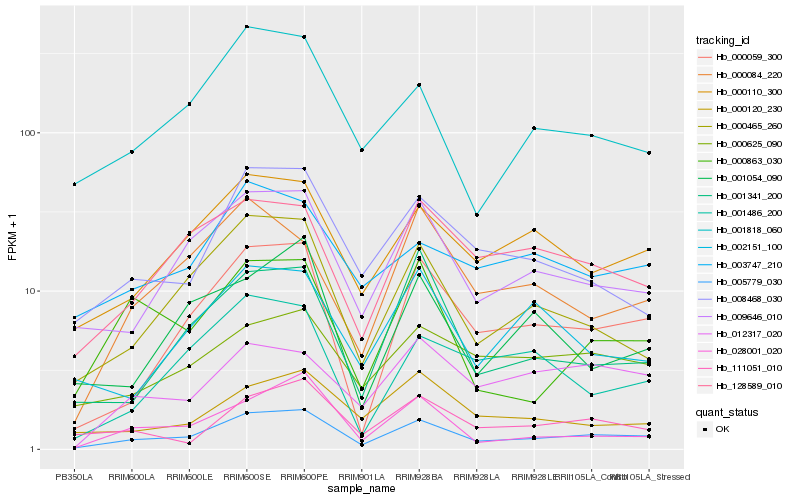
Hb_005779_030 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 2 |
Hb_009646_010 |
0.1264761791 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 3 |
Hb_000059_300 |
0.1279232565 |
- |
- |
caspase, putative [Ricinus communis] |
| 4 |
Hb_000110_300 |
0.1304656733 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 5 |
Hb_000465_260 |
0.1339193307 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 6 |
Hb_001818_060 |
0.1469990311 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 7 |
Hb_128589_010 |
0.1476400518 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 8 |
Hb_008468_030 |
0.150582738 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 9 |
Hb_000625_090 |
0.1539207609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_028001_020 |
0.1563699703 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 11 |
Hb_111051_010 |
0.160484305 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 12 |
Hb_003747_210 |
0.161818575 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 13 |
Hb_001341_200 |
0.1640482374 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 14 |
Hb_000120_230 |
0.1647515952 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 15 |
Hb_012317_020 |
0.1678874806 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_001486_200 |
0.1680577423 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 17 |
Hb_000863_030 |
0.1680616506 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 18 |
Hb_000084_220 |
0.1686507795 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 19 |
Hb_002151_100 |
0.1714408795 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 20 |
Hb_001054_090 |
0.1733388392 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |