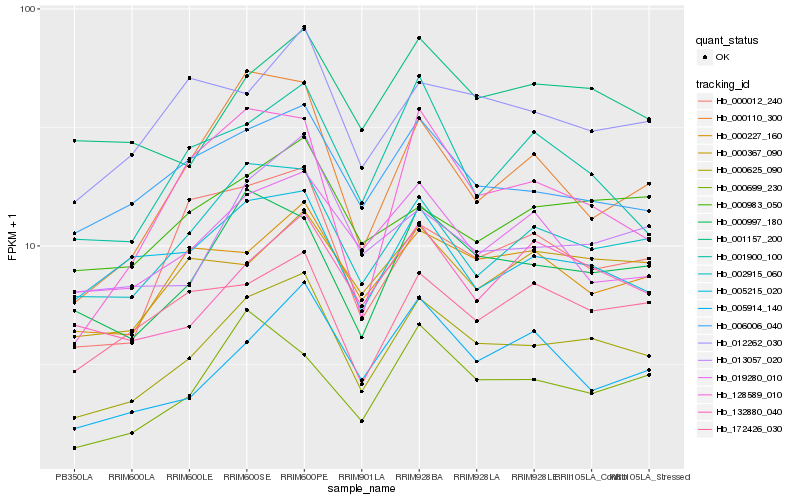
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000625_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002915_060 |
0.0931601548 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 3 |
Hb_000997_180 |
0.0955787017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_013057_020 |
0.1059913858 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 5 |
Hb_006006_040 |
0.1067816391 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 6 |
Hb_000227_160 |
0.1118579679 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 7 |
Hb_000110_300 |
0.1141502476 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 8 |
Hb_001157_200 |
0.1150313401 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 9 |
Hb_012262_030 |
0.1151067654 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000012_240 |
0.1168376888 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_172426_030 |
0.1173560792 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000983_050 |
0.1175398355 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_005914_140 |
0.117882425 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 14 |
Hb_019280_010 |
0.1183292414 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000367_090 |
0.1183299934 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 16 |
Hb_001900_100 |
0.1190024838 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 17 |
Hb_132880_040 |
0.119459429 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_128589_010 |
0.1205152524 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 19 |
Hb_000699_230 |
0.1207208642 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 20 |
Hb_005215_020 |
0.1219324048 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |