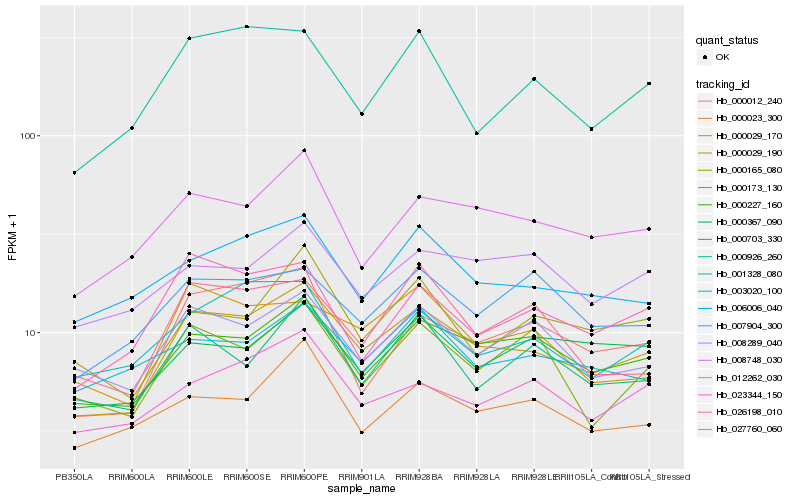
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_190 |
0.0 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 2 |
Hb_000227_160 |
0.0726152225 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 3 |
Hb_012262_030 |
0.0877798694 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_006006_040 |
0.0946608838 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 5 |
Hb_008289_040 |
0.0953384062 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 6 |
Hb_000012_240 |
0.1006686504 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_023344_150 |
0.1007491434 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 8 |
Hb_000926_260 |
0.1018422917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000703_330 |
0.1023942835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_026198_010 |
0.1024552268 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_300 |
0.1024815451 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 12 |
Hb_000029_170 |
0.1025691456 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 13 |
Hb_000165_080 |
0.1038154599 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007904_300 |
0.105226571 |
- |
- |
copine, putative [Ricinus communis] |
| 15 |
Hb_027760_060 |
0.105743581 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 16 |
Hb_001328_080 |
0.106024914 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 17 |
Hb_000173_130 |
0.1067910468 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003020_100 |
0.1070052479 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 19 |
Hb_008748_030 |
0.1073665456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000367_090 |
0.1078396077 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |