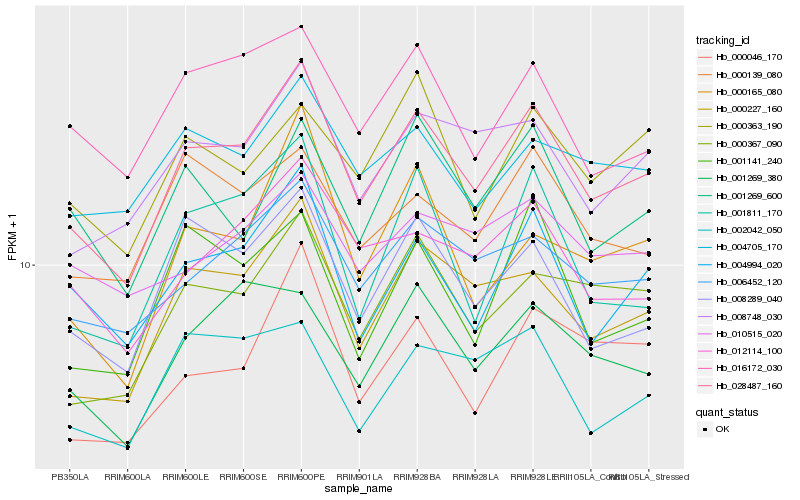
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028487_160 |
0.0 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 2 |
Hb_000227_160 |
0.058484875 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 3 |
Hb_006452_120 |
0.0638740637 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 4 |
Hb_008748_030 |
0.0688511274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004994_020 |
0.0688754992 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 6 |
Hb_000367_090 |
0.0691796525 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 7 |
Hb_008289_040 |
0.0702204397 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 8 |
Hb_012114_100 |
0.0727849833 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 9 |
Hb_016172_030 |
0.0775233044 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 10 |
Hb_001269_380 |
0.0781336167 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004705_170 |
0.0782101558 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 12 |
Hb_000139_080 |
0.0791994 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000046_170 |
0.080381842 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001269_600 |
0.0807854114 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_002042_050 |
0.0822127004 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000165_080 |
0.083266154 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001811_170 |
0.0835283951 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_001141_240 |
0.0843901776 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000363_190 |
0.0847733263 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_010515_020 |
0.0852362765 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |