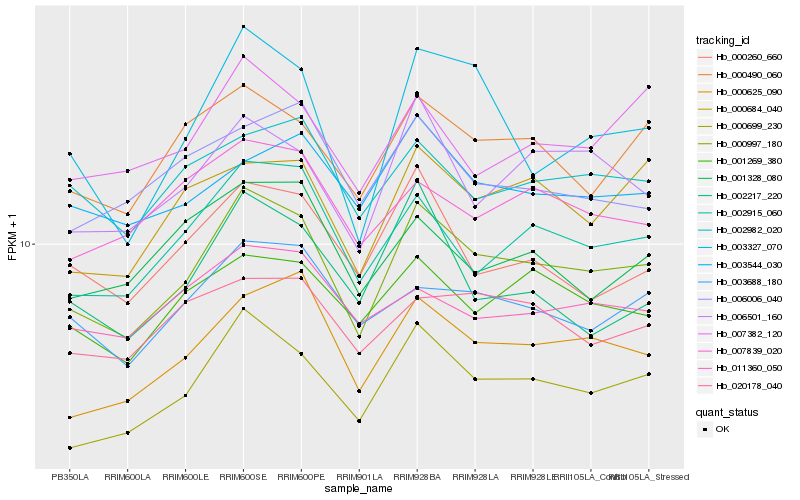
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000699_230 |
0.0835209823 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 3 |
Hb_002915_060 |
0.0918683657 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 4 |
Hb_000625_090 |
0.0955787017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003327_070 |
0.0997450182 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 6 |
Hb_000490_060 |
0.1009576007 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 7 |
Hb_006501_160 |
0.1016398799 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 8 |
Hb_001269_380 |
0.1071004581 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003688_180 |
0.1086966659 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 10 |
Hb_000684_040 |
0.1097061513 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 11 |
Hb_000260_660 |
0.1114979044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007382_120 |
0.1156008844 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_002982_020 |
0.1161758003 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 14 |
Hb_007839_020 |
0.1172603933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_020178_040 |
0.1177897596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003544_030 |
0.1200090339 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 17 |
Hb_006006_040 |
0.1216684951 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 18 |
Hb_001328_080 |
0.1218638318 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 19 |
Hb_011360_050 |
0.122195561 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002217_220 |
0.122242443 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |