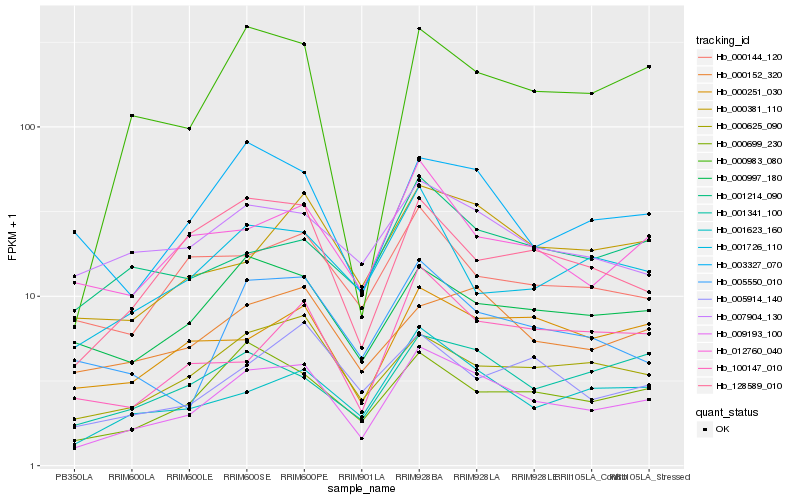
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009193_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 2 |
Hb_000699_230 |
0.1241036553 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 3 |
Hb_001623_160 |
0.1249123605 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 4 |
Hb_000625_090 |
0.1283649387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000381_110 |
0.1314259635 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000997_180 |
0.1344715149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000251_030 |
0.1371673714 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000152_320 |
0.1379920807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000983_080 |
0.1396906326 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 10 |
Hb_128589_010 |
0.1404498968 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 11 |
Hb_012760_040 |
0.1424469475 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 12 |
Hb_001341_100 |
0.1438190048 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 13 |
Hb_003327_070 |
0.1466911817 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 14 |
Hb_005550_010 |
0.1496632449 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 15 |
Hb_005914_140 |
0.1504993921 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 16 |
Hb_000144_120 |
0.1510453678 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_007904_130 |
0.1512955572 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 18 |
Hb_100147_010 |
0.1518092657 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 19 |
Hb_001214_090 |
0.1535342705 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 20 |
Hb_001726_110 |
0.1545450018 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |