| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
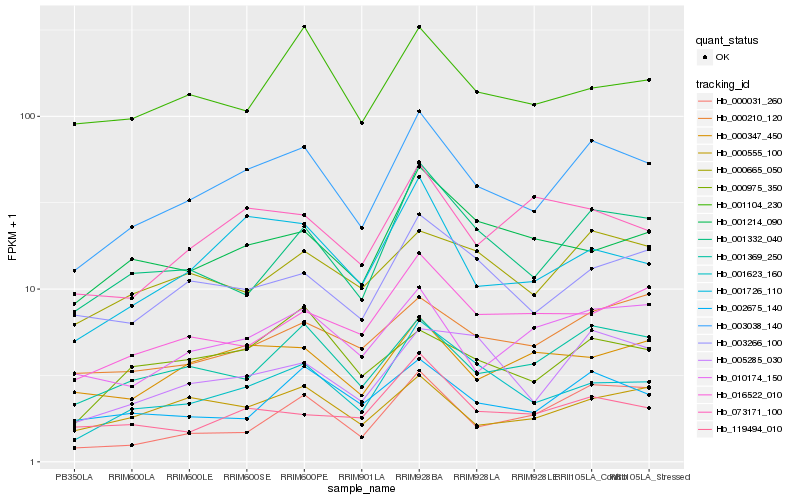
Hb_003038_140 |
0.0 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 2 |
Hb_001369_250 |
0.0995405711 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001726_110 |
0.1132483491 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 4 |
Hb_001623_160 |
0.1144751499 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 5 |
Hb_001332_040 |
0.1193867198 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 6 |
Hb_002675_140 |
0.1200746438 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 7 |
Hb_010174_150 |
0.1202540897 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_003266_100 |
0.1205754846 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 9 |
Hb_000665_050 |
0.1210061836 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 10 |
Hb_000555_100 |
0.1227831201 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 11 |
Hb_005285_030 |
0.1245080445 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 12 |
Hb_000975_350 |
0.1275729499 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 13 |
Hb_000347_450 |
0.1286057312 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000210_120 |
0.1288582842 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 15 |
Hb_073171_100 |
0.1345213274 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 16 |
Hb_016522_010 |
0.1350487916 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 17 |
Hb_001104_230 |
0.1372443551 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 18 |
Hb_119494_010 |
0.1380305601 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 19 |
Hb_001214_090 |
0.1384801303 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 20 |
Hb_000031_260 |
0.1389532283 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |