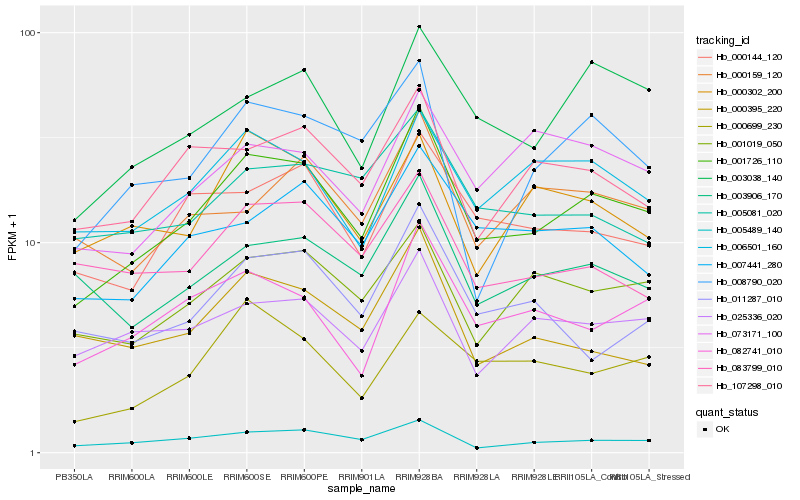
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001726_110 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 2 |
Hb_005489_140 |
0.1040840954 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 3 |
Hb_003038_140 |
0.1132483491 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 4 |
Hb_000144_120 |
0.1165362327 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_082741_010 |
0.1169198556 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 6 |
Hb_001019_050 |
0.1173245689 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 7 |
Hb_007441_280 |
0.1189095876 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 8 |
Hb_006501_160 |
0.1208340068 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 9 |
Hb_025336_020 |
0.1224794208 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 10 |
Hb_107298_010 |
0.123773475 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 11 |
Hb_003906_170 |
0.1244812586 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_008790_020 |
0.1288162042 |
- |
- |
PREDICTED: uncharacterized protein LOC105638122 [Jatropha curcas] |
| 13 |
Hb_000699_230 |
0.1303134937 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 14 |
Hb_005081_020 |
0.1308286043 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 15 |
Hb_073171_100 |
0.132415964 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 16 |
Hb_083799_010 |
0.1338346765 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000302_200 |
0.1347130613 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 18 |
Hb_000159_120 |
0.1349782616 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 19 |
Hb_011287_010 |
0.1350098641 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000395_220 |
0.1360506377 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |