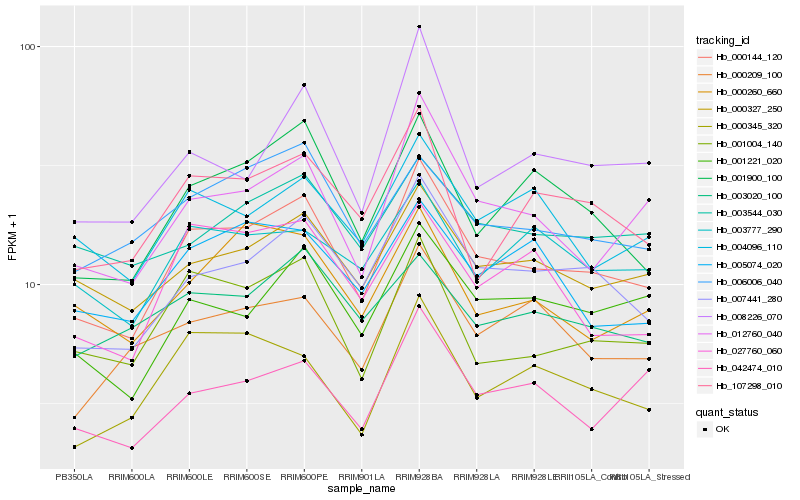
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000144_120 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_007441_280 |
0.0712744876 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 3 |
Hb_012760_040 |
0.0858830322 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 4 |
Hb_001004_140 |
0.0901156478 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 5 |
Hb_000327_250 |
0.0908851307 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 6 |
Hb_001900_100 |
0.0913665178 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001221_020 |
0.091397428 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 8 |
Hb_107298_010 |
0.1008120886 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 9 |
Hb_006006_040 |
0.101107333 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 10 |
Hb_027760_060 |
0.1028096724 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_000345_320 |
0.1040694606 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004096_110 |
0.1048723718 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 13 |
Hb_042474_010 |
0.1072219653 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 14 |
Hb_000260_660 |
0.1072257043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008226_070 |
0.1080905786 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 16 |
Hb_003777_290 |
0.1089501793 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 17 |
Hb_003544_030 |
0.1095719217 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 18 |
Hb_000209_100 |
0.1097974257 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003020_100 |
0.11004045 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 20 |
Hb_005074_020 |
0.1112200455 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |