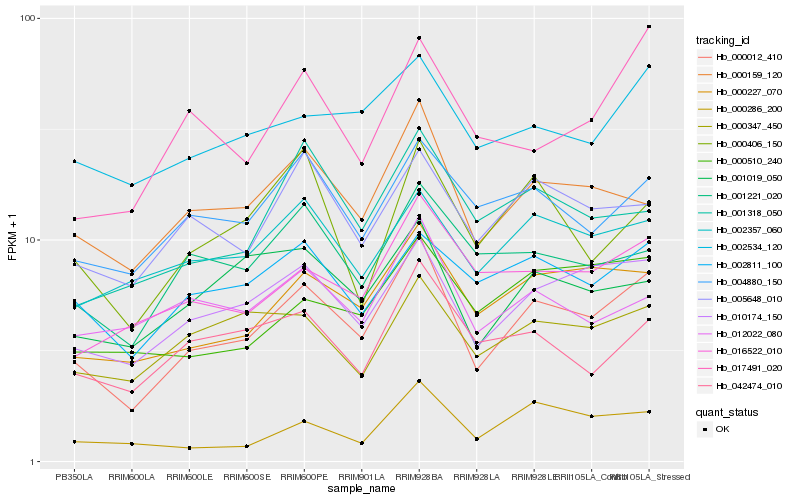
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_410 |
0.0 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_010174_150 |
0.0984949052 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000227_070 |
0.1078186216 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 4 |
Hb_000159_120 |
0.1184958372 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 5 |
Hb_016522_010 |
0.1314751435 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 6 |
Hb_004880_150 |
0.1327962041 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 7 |
Hb_002534_120 |
0.1329300334 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005648_010 |
0.1336077639 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 9 |
Hb_002811_100 |
0.1340258457 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 10 |
Hb_001019_050 |
0.1358338396 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 11 |
Hb_000347_450 |
0.1375679226 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002357_060 |
0.1378191771 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 13 |
Hb_000510_240 |
0.1380587935 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_001221_020 |
0.1394369279 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 15 |
Hb_042474_010 |
0.139637978 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 16 |
Hb_001318_050 |
0.1406887656 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 17 |
Hb_000286_200 |
0.1424977811 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000406_150 |
0.1433715859 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 19 |
Hb_012022_080 |
0.1441313585 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_017491_020 |
0.1448979693 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |