| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
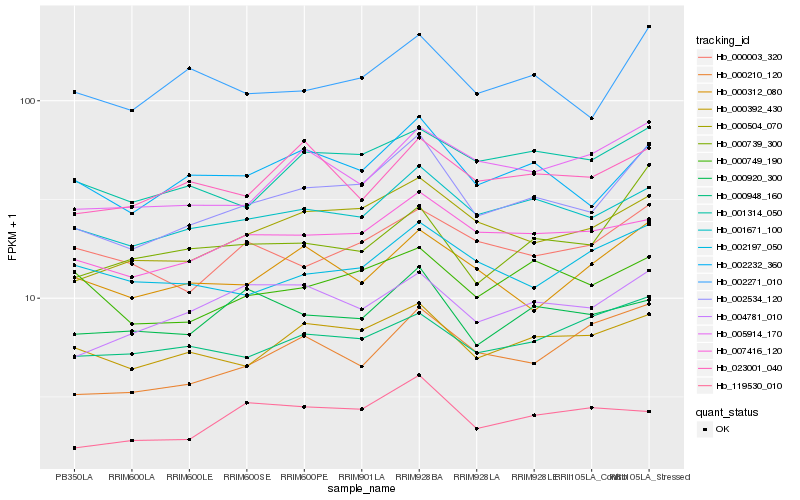
Hb_002534_120 |
0.0 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000504_070 |
0.0778439418 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_002232_360 |
0.0820307905 |
- |
- |
PREDICTED: proline iminopeptidase isoform X2 [Jatropha curcas] |
| 4 |
Hb_001671_100 |
0.0831797343 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 5 |
Hb_000392_430 |
0.088426756 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 6 |
Hb_002271_010 |
0.0899168978 |
- |
- |
PREDICTED: 60S ribosomal protein L8 [Jatropha curcas] |
| 7 |
Hb_007416_120 |
0.0915534821 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 8 |
Hb_001314_050 |
0.0952272182 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit alpha-like [Jatropha curcas] |
| 9 |
Hb_005914_170 |
0.0980010646 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4E-2-like [Gossypium raimondii] |
| 10 |
Hb_004781_010 |
0.1002041997 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 11 |
Hb_000210_120 |
0.1035290202 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000739_300 |
0.1044569729 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |
| 13 |
Hb_000003_320 |
0.1062664048 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 14 |
Hb_000749_190 |
0.1064088556 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Populus euphratica] |
| 15 |
Hb_000312_080 |
0.1070500381 |
- |
- |
PREDICTED: transmembrane protein 97-like [Jatropha curcas] |
| 16 |
Hb_000948_160 |
0.109214991 |
- |
- |
PREDICTED: 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_119530_010 |
0.1097586487 |
- |
- |
- |
| 18 |
Hb_023001_040 |
0.110719023 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000920_300 |
0.1110363781 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 20 |
Hb_002197_050 |
0.1112803583 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |