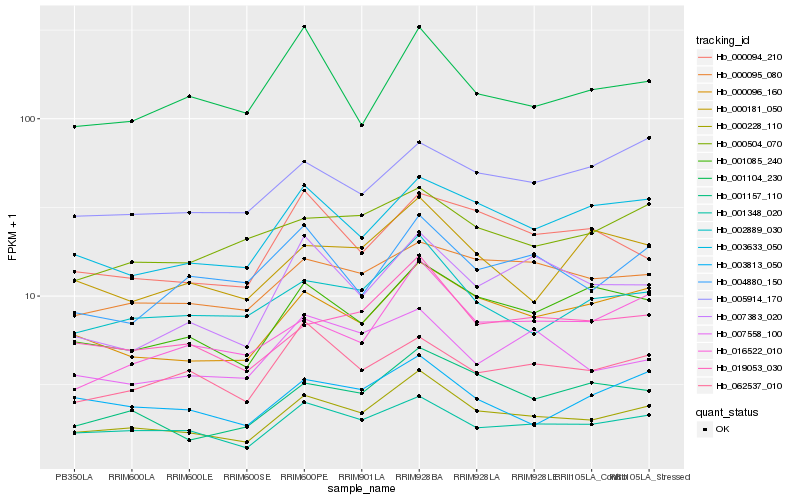
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000228_110 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_001348_020 |
0.0717352783 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000096_160 |
0.0761773584 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 4 |
Hb_019053_030 |
0.0794104996 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 5 |
Hb_001085_240 |
0.0859485859 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002889_030 |
0.0895400336 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 7 |
Hb_003633_050 |
0.0926310192 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 8 |
Hb_001157_110 |
0.0959666891 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 9 |
Hb_000095_080 |
0.0960739383 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 10 |
Hb_016522_010 |
0.1028295608 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 11 |
Hb_003813_050 |
0.1055714888 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_062537_010 |
0.108245533 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 13 |
Hb_000504_070 |
0.1091039435 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_007558_100 |
0.1094699031 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000094_210 |
0.1100271754 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 16 |
Hb_000181_050 |
0.1105818515 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 17 |
Hb_007383_020 |
0.1119016517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005914_170 |
0.1121745078 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4E-2-like [Gossypium raimondii] |
| 19 |
Hb_001104_230 |
0.1147859105 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 20 |
Hb_004880_150 |
0.1150072706 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |