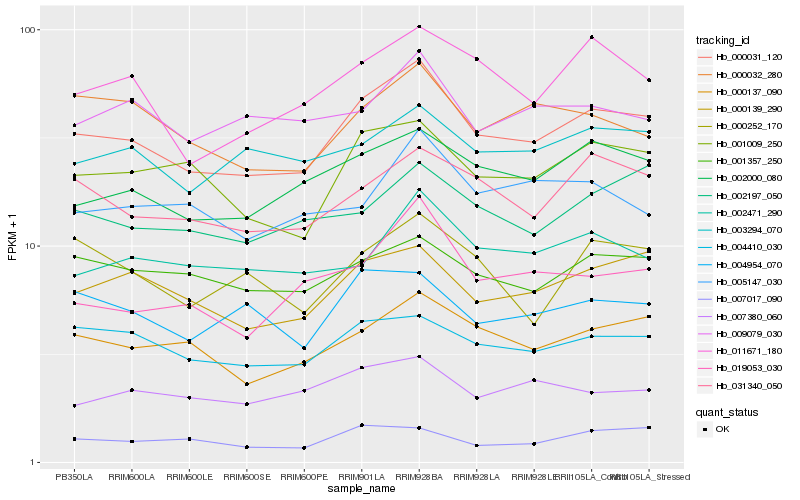
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 2 |
Hb_000137_090 |
0.0776148153 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Vitis vinifera] |
| 3 |
Hb_002000_080 |
0.0791184987 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000139_290 |
0.0806549906 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001009_250 |
0.0814576677 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_031340_050 |
0.0843815333 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_011671_180 |
0.0866331674 |
- |
- |
PREDICTED: probable quinone oxidoreductase [Jatropha curcas] |
| 8 |
Hb_004410_030 |
0.0879556637 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_019053_030 |
0.094193561 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 10 |
Hb_002471_290 |
0.094893828 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_001357_250 |
0.0955541705 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 12 |
Hb_003294_070 |
0.0957803634 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 13 |
Hb_004954_070 |
0.0968387646 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 14 |
Hb_000032_280 |
0.0969420307 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 15 |
Hb_002197_050 |
0.0990055125 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 16 |
Hb_007380_060 |
0.1001357358 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |
| 17 |
Hb_007017_090 |
0.1003526796 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |
| 18 |
Hb_009079_030 |
0.1012613174 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 19 |
Hb_005147_030 |
0.1014701779 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 20 |
Hb_000252_170 |
0.1026972216 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |